|
Inferring the Past
© by
Paul Handford
|
Zoology Department
University of Western Ontario
LONDON, Canada, N6A 5B7
handford@julian.uwo.ca
|
This is an english-language version of
a paper invited by Ciencia Hoy, which is a sort of Argentine Scientific
American.
It is due to be published in August 1999.
The living world has impressed humanity from our earliest days, and
its exuberant diversity and remarkable fit to the varied conditions of
life have always challenged our understanding. Expanding exploration
of the world and its contents has led to the realization that nature not
only is astonishingly diverse and "well-designed," but also shows other
marked characteristics, notably a clear "hierarchical order" in the levels
of resemblance among organisms, and an irregular distribution of their
kinds about the globe (no tigers in South America, no kangaroos in North
America, no lemurs in New Guinea, and suchlike.) And there are also
fossils - ancient life, often radically different from modern forms and
now extinct. Eventually, since the mid-19th century, our earlier
explanations of these phenomena through providential creation and other
supernatural agencies have given way to the view that life's characteristics
could be rendered intelligible in a fashion acceptable to modern science
by the notion of evolutionary descent and change
One of the keys to the development of the notion of evolutionary change
and diversification of the world's biota was the discovery and deployment
of the notion of deep time: the earth has a very long history.
The Earth and its life did not suddenly spring into existence; rather
they developed, across an immense expanse of time. Thus,
so we now understand, today's organisms descended through myriad generations,
slowly and severally spreading across the globe as circumstances permitted,
adapting and diversifying as they went. This long historical narrative
of the biota, from its most distant and simple origins at least 3.8 billion
years ago, through prokaryote (bacterial) forms, to unicellular eukaryotes
around 1.6 billion years ago, and to multicellular forms such as algae,
fungi, plants and animals in the last few hundred million years,
has taken place against a backdrop of slowly shifting global geography
and chemistry.
This is the standard "big picture" narrative of the evolutionary development
of the earth's biota. But how do we know what the details of such
narrative histories are? Having come to understand that all life
is connected in a vast ramifying genealogical thicket, extending through
nearly 4 billion years, how do we know which forms are related to which,
and how closely? In short, how do we reconstruct life's evolutionary
past?
The Problem: Ambiguity
in the clues to history
The problem of working out what has actually happened in the past is
a very general one. It ranges from the cosmic scale - when &
how did the universe begin, and what happened to it along the way to the
present? - to the local scale - how did John F. Kennedy die, who
killed him, how, and why? In between these extremes lie such matters
as: the development of the world's atmosphere, continents and oceans,
the evolutionary descent of the world's organisms, the history of humans
and their cultures & languages, and the political, social and economic
history of peoples and nations. In approaching an answer to any such
historical problem, we must carefully scrutinize whatever it is that may
carry information about the past through time. We must be very clear
about exactly where such information may lie, how it should be interpreted,
and how the information may become distorted. Here I wish to make
some observations about difficulties that arise when we address this biological
problem: how can we reconstruct
life's evolutionary genealogy (1)?
Where are the clues to this history? What are the difficulties in
their decipherment? The answers to such questions have repercussions
in many areas of evolutionary biology and they have, accordingly, been
approached many times, indeed they constitute classic issues within evolutionary
science. As our knowledge has progressed, we have discovered that
the problem is ever more subtle.
Charles Darwin and Alfred Wallace, as well as persuading us that the
biota, of which we are a part, has changed and diversified, also suggested
to us a mechanism by which this change might come about: given organic
multiplication, heritable variation in attributes that affect that multiplication,
and a finite world of resources, then, as conditions change through time,
so too must the characteristics shown by lineages of organisms - they will
change, and do so non-randomly (adaptively) with respect to those changing
conditions. This process is called natural
selection. Other changes prosper solely, or largely, by chance,
that is, they have no bearing upon the rate of multiplication, and this
process in known as genetic drift. As time passes, character changes
accumulate. This view of things is termed "descent with modification"
- organisms inherit most of their structure, unchanged, from their ancestors,
but bits of that structure change over the generations, and those changes
accumulate. This means that, in general, the more closely related organisms
are (that is, the closer to the present time their most recent common ancestor
lies) the more closely they will resemble one another, simply because there
will have been less time for heritable changes to accumulate. This
is Darwin's explanation for the long-recognised hierarchical system of
resemblance among organisms alluded to in the first paragraph. Such
resemblance will be reflected in many aspects of structure, from external
anatomy, through details of internal & cellular anatomy, to the structure
of the hereditary molecules themselves - DNA and RNA. It is this
similarity system which constitutes the text in which are written all the
available clues to the history of life.
But there is a difficulty: it is not altogether clear how "structural
similarity" should be interpreted in any given case, for the simple reason
that organisms may resemble one another for reasons besides propinquity
of evolutionary descent. Resemblances may reflect the particular
actions of natural selection independently of any particular level of relatedness
(recency of common ancestry): they may represent similar adaptive
responses to similar environmental exigencies. Evolutionary change
may not always be simply divergent. Resemblances may also arise by
chance. Similarities due to causes other than close descent relationship
are termed homoplastic or convergent. Thus we have a genuine problem
of historical inference - what do characters mean? what information
do they carry? - information about propinquity of descent (history)?
information about similarity of adaptation to conditions (natural selection)?
or information about the vagaries of chance? A simple illustration
of the nature of this problem is provided by a case where a known history
can be compared with that which might be inferred from characters of the
sort routinely used for assessing historical (phylogenetic) relationships.
Domestic pigeons are derived from the wild Eurasian Rock Dove, Columba
livia. We have good evidence that this domestication process
began at least 5000 years ago. At various times and places, domesticated
pigeons have escaped from captivity and returned to the wild, forming populations
of feral birds. This process began in Europe shortly after initial
domestication. The wild Rock Dove does not exist in North America,
but feral populations do, and they have derived, no more than 400 years
ago, from domesticated stocks brought over by Europeans. We
may use the morphology of wild Columba livia, ancient European
and recent North American lineages of feral birds, and domestic stocks
to infer their evolutionary relationships; this inference may be
compared with their known historical relationships. This comparison
has been made by Richard Johnston (2),
and the result of interest here is shown in Fig.1.
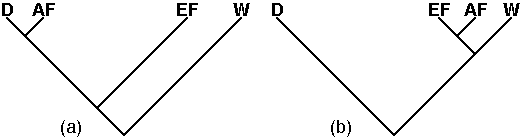
|
| Fig. 1 The historical relationships
among: wild Columba livia (W), domestic pigeons (D), and feral
pigeons from Europe (EF) and from North America (AF), as known from archeological
& historical evidence (a), and as implied by morphological resemblances
(b). It is clear that the similarities in morphological characters
provide a poor estimate of the genealogical reality. |
Morphologically, the feral populations closely resemble each other and
wild Columba livia, rather than the domestic stocks to which they
are most closely related in time. It is evident that the discrepancy
between the history inferred from morphological characters and the known
history has come about because natural selection has modified wild-living
birds, whether feral or original Columba livia, to conform to a
particular morphology, regardless of evolutionary origins. Despite
the great morphological similarities in the characters shown by ferals
and wild doves, they are not aquired by close inheritance, but through
a round-about route of evolutionary convergence. Despite their very
high level of similarity, the characters are not strictly "the same";
they are functional analogues, not true homologues, underlain by the same
genome. They mislead us about history.
This is the same sort of phenomenon that occurs in selection experiments
on replicate lineages of micro-organisms. Lineages that are selected
to grow on the same novel medium may all become adapted to that medium
- they will achieve the "same" phenotype at one level of analysis (capacity
to grow in the medium), but they will often achieve this adaptation in
different ways, which will be "recorded" differently in their genomes since
they derive from different mutations. After selection, the adaptive
phenotypes of the lineages will tell us nothing about their historical
relationships with one another (or with other strains perhaps involved
in other experiments selecting for other capacities.) Genotypes continually
diverge, tracking history, while simultaneously, phenotypes may diverge,
remain largely the same, or converge, as circumstances dictate.
Some recent work by Michael Travisano and others emphasises this repeatability
of phenotypic evolution.
This discussion brings into focus a central problem in evolutionary
biology: that of the distinction of analogous from homologous
characters. The confusion of analogy and homology, and our
struggles to unravel them, have a very long history in biology. It
is the confusion that lay behind our once classifying whales as fish.
One of the points of this essay is that our struggles are far from over.
Even the meaning of homology itself is not clear - large quantities of
ink have been spent upon the discussion of its slippery
nature (2a). We may
note the following attitudes as indicating the range of emphasis:
-
Owen 1843. "the same organ in different animals under every variety
of form and function." Here, the issue is one of essential similarity
in structure. Later, evolutionists (e.g. Lankester) added common
ancestry to an otherwise similar definition.
-
Remane 1952. Evolutionary homology involves similarity in relative
position, in structural detail, and the presence of transitional forms
(including those in ontogeny.)
-
Sneath & Sokal 1973. Characters are homologous if they are "very
much alike in general and particular."
-
Patterson 1982. Homology equates to synapomorphy [a derived trait
characterizing a monophyletic group]. Thus homology is directly tied
to common descent.
-
Van Valen 1982. Homology is "a correspondence between two or more
characteristics of organisms that is caused by continuity of information."
-
Roth 1984. Homology is "based on the sharing of pathways of development
which are controlled by genealogically-related genes."
-
Raff 1996. Homologous features are "those that share a common
evolutionary origin and an underlying commonality of morphology, resulting
from a continuity of information."
We may see in this diverse array of attitudes a distinction between lineages
(which are unique) and the (perhaps not-so-unique) characters
which typify them. There are implicit questions here: how should
a character be delineated? what is a character?
-simply "a structure"? the structure and its detailed relationships
with other structures? or the structure and its unique genetic/environmental
interactive recipe? This is a serious problem, indeed. We now
know that there is no straightforward mapping of genes, or of the developmental
pathways guided by them, onto structural characters. Whale and fish
fins may now be readily recognised by close inspection as analogous structures,
but what about dove bones of different shapes & sizes?
Modern phylogenetic analytic practice makes much use of the principle
of parsimony (3), both in
the reconstruction of evolutionary trees, and in the establishment of homologies.
However, there are real difficulties attached to the application of these
techniques to morphological characters, for they are ultimately based in
assumptions about the commonness of convergence and parallelism, and about
the commonness, speed and reversibility of evolutionary character change,
which is very much begging the question here.
Carrol & Dong (4) express
this problem as follows: "Is there any category of characters that
is not commonly subject to convergence? Is it ever possible to establish
phylogenetic relationships without some specific information regarding
the strict homology of the characters in question?.......... The principle
of parsimony cannot be used directly to identify homologous characters
if most of the derived characters are convergent." The problem therefore
seems to be one of: at what level should we scrutinise the
organisms? The further we get away from the genome or genotype, the
more prey we are to being misled by adaptive changes, and by complications
from "gene-character" relationships in the developmental system; structural
characters necessarily retain some unknowable degree of ambiguity.
So we are not yet of one mind on what homology is, what it ought to
be, or how to recognise it in practice. However, we can see some
developing focus, in the list of quotations above, on the idea that it
should make some explicit reference to the continuity of genetic information,
and not simply to the apparent nature of the characters themselves.
Further, even if we settle on any one of the definitions, we still find
it very difficult to recognise with real security true inherited homologies
in the "real world." Our new understanding of the complex involvements
of genes, developmental pathways, and body characters makes it much more
difficult to decipher the historical meaning of structure.
This seems to be the clear message coming from the selection experiments
on bacteria alluded to earlier.
Where then may we find reliable
information about history?
Organisms reproduce by transferring information about their own body
construction across generational time. The mechanisms for such information-transmission
constitute inheritance. Evolutionary descent involves the
inheritance of (slightly altered) information from generation to generation
where it is translated, and re-translated each generation, into new bodily
forms through the mechanisms of development. To permit
secure inferences about descent relationships, then, we must gain access
to the information which is passed among the generations (the genetic material
- what we might call the "primary text" of phyletic history), or
decipher it, if we can, from the structures which this information generates
through development (aspects of organisms' bodies - the "developmentally-encrypted
text."). Until very recently, we have only had access to the
"encrypted text" - virtually the entire edifice of phylogenetic science
has been erected on a base provided by what we have thought we could decipher
of genealogy's primary text through the variously-refracting prism of development
from genome to phenotype. It is but equally recently that we have
begun to recognise just how distorting that prism can be. We have
traditionally thought of the genome's evolutionary changes largely in terms
of the expansion and replacement of genetic instructions for building bodies
- of the organisms' developmental "recipes" becoming slowly overwritten
into something else. But now we know that things are much more complicated:
genomes in fact retain vast archives of "code," some from the far-distant
evolutionary past, though the characters that this code once produced may
be long gone from the lineage (as an example, chicken embryos can be induced
to produce tooth buds (5)),
or the code may now be serving functions in entirely different developmental
contexts, producing entirely different characters (examples of this are
now well known since the discovery and understanding of homologous homeobox
genes in a wide array of animal forms (6).)
This clearly has enormous repercussions on the uniqueness of characters
& change in evolutionary lineages.
For these reasons, it seems that we would be well-advised to address
our inquiries about the past directly to the primary text, since there
is an inevitable ambiguity in those characters which are generated (or
not generated) from the developmental translation of that text into bodily
structures. It must certainly be admitted that there are some difficulties
in the interpretation of similarities at the genomic level too; nonetheless,
we have some good a priori reasons, hinted at above, for supposing that
scrutiny of the molecules of heredity will help us reliably to read history,
and recent empirical studies endorse
this view (7).
In most cases, phylogenies generated by analyses of molecular data support
a great deal of the picture developed from analyses of classic morphological
characters. On the other hand, molecular phylogenies sometimes contradict
traditional analyses, providing surprises; sometimes these surprises
are substantial. Wherever we find surprises, this means that we have
uncovered potential cases of previously unrecognised evolutionary convergence
- we have uncovered cases where history has been erased from organismal
structure.
Surprises from Molecular Phylogenetics
Let us now look at some examples which together demonstrate that not
only can morphological characters mislead us about history, but that such
distractions can generate mistaken taxonomies
and classifications (8), sometimes
on a massive scale, and that evolutionary processes can repeatedly produce
near-identical forms from disparate origins. I shall eventually conclude
that we have good prima facie evidence that, at the level of the phenotype,
far from being seriously compromised and constrained by contingent history,
large-scale evolutionary narratives may well have a strong predictable
element - that the major features of earth's biota would probably be much
the same, in phenotypic terms, no matter how often the experiment
be run (9).
False cousins within the family.
The doves considered earlier, whether wild, feral or domesticated, are
all inter-fertile - are all part of the same biological species.
But can mistaken inferences about historical relationships involve forms
that belong to different species? I shall briefly describe three examples
which show that the answer seems to be yes.
First consider an example derived from some of our ongoing studies of
the south American passerine bird fauna, in this case involving the widespread
genus Poospiza
- the warbling finches (10).
These small birds are quite diverse, and range from sea-level to over 4000m,
mainly in scrubby habitats south of the equator. An hypothetical
phylogeny for the seventeen species has recently been proposed, based in
morphology, size, coloration and geographical range, by the Danish ornithologist Jon
Fjeldså (11).
Our work comprises a phylogenetic analysis based on variation in a 1324-base
sequence from two mitochondrial genes, 16S RNA and cytochrome B.
This latter gene has been used extensively in phylogenetic analyses of
birds and other taxa, and is widely regarded as providing a reliable phylogenetic
signal. In our results, we find very little support for Fjeldså's
picture (see Fig. 2).
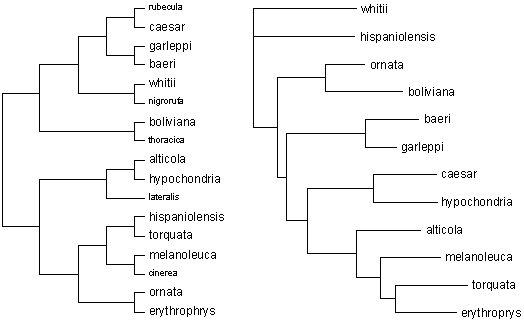
|
| Figure 2. Phylogenetic
relationships among the species of Poospiza as hypothesised (left)
by Fjeldså, on the basis of morphology, plumage, behaviour and geographic
range, and (right) by Lougheed & Handford, on a basis of mitochondrial
DNA sequence variation. Fjeldså's study involves all of the
species within the genus, while the 5 taxa shown in small type were not
included in the DNA analysis. Note that the two hypotheses concur
only in the close relationship among baeri and garleppi. See text
for other commentary. |
We may see that only one (baeri-garleppi) of Fjeldså's
sister-taxon pairs is recovered in the DNA analysis. The differences
between the two phylogenetic hypotheses include: 1. our very deep separation
between torquata and hispaniolensis, and between ornata and
erythrophrys, (which pairs are morphologically very similar indeed)
and 2. our finding of strong associations of: a) caesar
with hypochondria, b) alticola with melanoleuca,
c) torquata and erythrophrys, and d) boliviana
with ornata. This study strongly suggests that the kinds
of morphological differences that exist among members of this genus are
a very poor guide to evolutionary history.
But what of more substantial differences, such as exist among members
of a whole family? In a study conducted by Daniel
Ellsworth and colleagues (12)
of the historical evolutionary relationships among all of the North American
grouse (family Tetraonide), using variation in mitochondrial DNA sequences,
it was found that, in the main, the current understanding of relationships
was endorsed; however the two members of the genus Dendragapus,
D. obscurus and D. canadensis, are, according to the molecular
phylogeny, not each other's closest relative (as should be the case, since
they are in the same genus.) Instead, the Spruce Grouse (D. canadensis)
is allied with the Ruffed Grouse (Bonasa umbellus) and the Sage
Grouse (Centrocercus urophasianus), while the Blue Grouse (D.
obscurus) is involved with the ptarmigan (Lagopus spp.) and
the capercaillie (Tetrao urogallus), a Eurasian forest grouse (see
Figure 3).
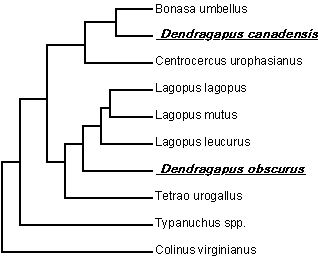
|
| Figure 3. Phylogenetic hypothesis
for North American members of the Tetraonidae, derived from analysis of
mitochondrial DNA sequence data. Evident is the disparate placement
of the two members of the genus Dendragapus. Colinus virginianus
is a quail (Phasianidae), used as an outgroup to root the tree. |
This phylogeny may be interpreted as suggesting that D. obscurus
derives from an Old World lineage that entered North America via Arctic
latitudes, while D. canadensis derives from a longer-established,
more southerly, North American lineage. It appears, then, that the
similarities that the "Dendragapus" species bear to one another, sufficient
to persuade taxonomists of their close evolutionary affinity, have been
independently derived, and possibly have something to do with adaptation
to their similar habitats in the coniferous forests of northern & western
N. America.
The New World, from Canada to Patagonia, is home to a large and very
diverse subfamily of songbirds, the Icterinae, which includes such Argentine
species as blackbirds, cowbirds, caciques and meadowlarks. Prominent
among these is the widespread genus Agelaius, which includes 9 species
(4 in Argentina). A recent study by Lanyon
(13) has demonstrated that
this genus is not, as evolutionary biologists put it, a "natural taxon."
That is, its member species are not all each other's closest relatives
- exactly as was the case in the grouse genus Dendragapus.
It now seems clear that the north and south American "Agelaius" species
have closer relatives among other members of the icterine avifauna of the
two respective continents: that is, the south and north American
forms are independently derived. In this case then, the morphology
of an "Agelaius" has been repeatedly evolved from different lineages spread
over two continents.
Striking and Global-level surprises
Fine examples of just how misleading morphology can be is provided by
the work of Sibley & Ahlquist
(14). These pioneers
of the molecular taxonomy of birds have amassed an enormous database of
among-taxon distance measures based in the technique of DNA-DNA hybridization
and, although their hypothetical phylogenies and classifications endorse
in large part the traditional classifications based in comparative anatomy,
they have provided some real surprises. Several avian taxonomists have
argued against many of their claims about relationships, but over the last
several years, independent work (15)
has shown that much of the Sibley & Ahlquist picture has substance.
Here I shall mention only two of their striking cases, ones which demonstrate
the enormous power of natural selection to mould anatomical characters
and so mislead us about history.
The New World has several species of carrion-eating bird known as condors
and vultures (Vultur, Sarcoramphus, Cathartes, Coragyps and Gymnogyps),
and they are classified into their own family of Cathartidae, distinct
from the Old World vultures, which they strongly resemble in many respects,
who are in turn members of the family Accipitridae (which also contains
the widely-distributed eagles and hawks). Together with the falcons,
all these birds are collected into the Order Accipitriformes, the birds
of prey. Thus, traditional taxonomy recognises that New and
Old World vultures represent parallel forms, from not very distinct origins,
within a single order of birds. Sibley and Ahlquist startled the
ornithological world when they claimed, based in their DNA-hybridization
work, that cathartids and Old World vultures are instead amazingly convergent
forms, from highly divergent lineages: cathartids, they claimed,
are members of (find their closest relatives among) the group including
storks and pelicans! Part of their phylogenetic conclusions are illustrated
in Fig. 4. These conclusions, which are now generally accepted,
concur with several other independent lines of evidence, both behavioural
and morphological, which have brought other ornithologists to suggest the
same relationships among these birds.
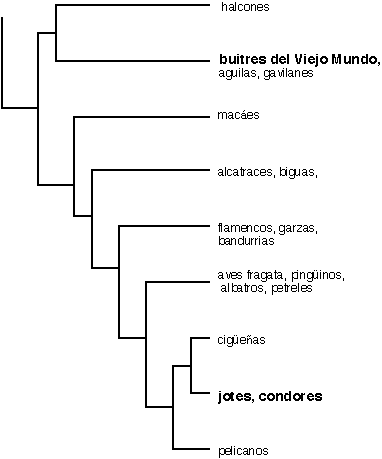
|
| Figure 4. Simplified version
of part of the Sibley & Ahlquist phylogeny of birds, based on DNA-DNA
hybridization measures, emphasising the relative position of the Old World
vultures (buitres del Viejo Mundo), as part of the family Accipitridae,
including eagles (aguilas), harriers (gavilanes) and hawks, and their New
World ecological counterparts, the highly convergent Cathartidae (jotes,
condores), as close relatives of storks (ciguenas) and pelicans (pelicanos).
Halcones = falcons; macaes = grebes; alcatraces = gannets + boobies; biguas
= cormorants; flamencos = flamingos; garzas = egrets + herons; bandurrias
= ibises; aves fragata = frigate birds. |
Another part of Sibley & Ahlquist's work has provided a stunning
resolution to a long-standing biogeographical puzzle - the "missing" Australasian
avifauna. It is well-known that, for somewhere in excess of 60 million
years, South America and Australia were island continents. Australia
remains an island, while South America ceased its island status a mere
3-5 million years ago. During all this time of isolation, occupying
much of the Cenozoic era, their biotas evolved independently of that of
the rest of the world, and this is eloquently reflected by the highly distinctive
nature of their mammal faunas, both living and extinct, where marsupials
are prominent or overwhelmingly dominant. We would expect that this
long isolation would also be evident in the avifauna, since birds also
evolved explosively during the Cenozoic era. It is indeed the case
that South America is possessed of a highly-distinct bird fauna, most notably
in housing all but about 50 of the world's suboscine passerines, a group
that contains over a thousand species, from woodcreepers and ovenbirds
to antbirds, tapaculos and cotingas, to flycatchers and kingbirds.
However, while Australia does indeed have many highly distinctive forms,
such as the lyre bird, the great majority of its avifauna is referred to
groups well-represented in Asia and elsewhere in the Old World. Sibley
& Ahlqist's study of the Australasian avifauna has revolutionised our
understanding of its evolutionary and biogeographic status. Far from
belonging primarily to other Old World lineages, Sibley & Ahlqist's
analyses reveal that the great mass of the Australasian avifauna is autochthonous:
that virtually all of its members are more closely related to each other
than any is to groups outside Australasia. What this means is that
this independently-evolving avifauna has generated forms so strongly convergent
on other Old World forms as to result in their being referred to entirely
mistaken groups. So the Australasian avifauna is not missing at all
- it is simply so well adapted to conditions similar to those elsewhere
in the world as to have almost completely erased its own evolutionary history
- its own genealogy - as construed from the perspective of anatomy.
These examples demonstrate clearly that the more effective natural selection
is in moulding the appearance of organisms' structure so as to reflect
their environmental circumstances, generating very closely-similar characters
in organisms of different phyletic lineages, the more likely it is that
these similarities among taxa will be seen as homologies, and therefore
ascribed to common ancestry, the true phylogeny be missed, and the selection
go unrecognised. Particularly effective selection leads to selection's
actions going unrecognised! How powerful can selection actually
be in erasing the historical message encrypted in organismal structure?
It is not possible to make any definitive statement on this matter at present,
but it is possible to use the historical record of systematics to demonstrate
that selection has often been sufficiently powerful at the level of the
external phenotype as to lead systematists into serious errors regarding
homology and, thereby, regarding history and comprehension of the true
face of evolutionary change. My inference from such cases is that
phenotypic evolution, through its sensitivity to natural selection, has
commonly provided patterns which has misinformed us about history;
that many features of the biota are more predictable than current convention
would have it; and that, if the "tape of life on earth" could be
re-run many times, then, provided that earth's physical and chemical conditions
are reproduced, so would the history of life's phenotypes, albeit involving
different, contingent, genomes (lineages).
Morphological evolution can repeat
itself
The cases described above, together with a growing number of others,
provide good evidence that even today we continue to be distracted by similarity
in morphological characters into mistaken inferences about evolutionary
history. In the terms that I introduced earlier, the developmentally-encrypted
text turns out to have been in a code that we failed to break and interpret
correctly; such message as there was in anatomy had since been modified
by natural selection beyond all recognition. This conclusion became
possible after analysis of the primary text - the genome. In addition
we have seen in several of these instances, and at various taxonomic levels,
that suites of characters have been generated several, even many, times.
This bears on the issues of the nature of characters and their interactions
with evolutionary lineages, and of the repeatability, even predictability,
of evolutionary change.
Finally, I shall describe a very recent and powerfully compelling example
illustrative of just these last matters - how evolutionary radiations in
similar environments can overcome history and repeatedly produce remarkably
similar evolutionary outcomes. The case is that provided by the work
of Losos and colleagues (16)
on Anolis lizard species of the Greater Antilles. Anoles are
major elements of the Caribbean biota, and on each of the Greater Antilles
(Cuba, Hispaniola, Jamaica & Puerto Rico) lizard assemblages include
similar arrays of species specialized to use the same array of microhabitats.
Specialization on these microhabitats has generated morphologies which
are highly repeatable from island to island, such that lizard species from
any given habitat on all islands resemble one another far more than they
do other lizard species from their own island. Did these various
habitat-specialist lizards each evolve once and then colonise the various
islands? or did ancestral lizards colonize the islands and independently
differentiate into the local habitat-related forms? A phylogeny reconstructed
using morphological characters overwhelmingly suggests the former scenario;
this is because algorithms for reconstructing phylogenies usually incorporate
an assumption of parsimony - that evolutionary change is minimal, meaning
that repeated evolution of the "same" character is regarded as rare or
absent. While this may well be true for molecular characters over
relatively short time-spans, we have swiftly-growing evidence suggesting
that this is not the case with morphological characters. As it turns
out, a phylogenetic analysis of the anole species based on mitochondrial
DNA sequence variation powerfully rejects this picture from morphology,
and instead shows that, with only two exceptions, similar lizards from
different islands are not closely related. In contrast, it appears
that there has been a large number of evolutionary character transitions,
repeatedly generating "the same" characters in different evolutionary lineages,
and in different historical sequence. One might wonder whether Anolis
lizards can only be represented by a few forms, and thus their repeated
evolution not surprising; the existence of a much wider array of
taxa elsewhere in the Caribbean region makes this possibility very unlikely.
Knowability vs. Predictability;
Characters vs. Lineages.
We began with questions concerning the knowability of the past:
how we might infer historical descent relations - how we might trace lineages
through time. It has been argued above that we should attend closely
to the messages in the primary text of the genome and be wary of the distractions
provided by characters with a strong potential for ambiguity generated
by embryological processes (morphology). But in considering the illustrative
examples, we have come also to talk of predictability. We need to
discuss these two concepts together, and clearly distinguish them.
The knowability of the past depends upon the processes linking
that past with the present: what do these processes do to information
about the past? do they tend to preserve it? or do they tend
to destroy it? Though related to it, this question is
distinct, in a very important way, from: Is the present predictable
from the past? This distinction arises because the processes involved
in changing aspects of organisms through time may be either directed (such
as by natural selection) or undirected (such as the drift in the frequency
of neutral mutations), and these processes differ substantially in their
capacity to leave lasting traces in which history may be read. Structures
primarily affected by undirected processes (like much molecular sequence
structure) can retain long-lasting traces which permit discovery of the
past, but not allow prediction or retrodiction - since much of the path
is random. On the other hand, selection will tend to erase traces
of the past in characters which are associated with fitness (like most
morphological characters), so the discovery of the specific path from initial
morphological conditions to eventual outcomes may be lost; however,
at the level of the adaptive characters themselves, some prediction is
possible. Characters therefore differ substantially in their capacity
to provide knowledge of the past: the more they are subject to selection,
the less likely they are to provide secure historical information.
It is for this reason that taxonomists have traditionally pursued that
elusive entity "the non-adaptive character set". Unfortunately, such
designations often rest upon our ignorance of the significance of characters
in the life of the organisms, and assertions about functionality
(17). Clearly,
the structural characters assayed in the dove example above had been subject
to processes which obscured their lineage; nevertheless, the form
of those characters is, in principle, predictable from adequate knowledge
of the conditions of life in the wild as opposed to life in captivity.
To dramatise the difference between knowability and predictability,
we might ask: given some window onto the information-content of relevant
extinct and extant organisms, could we discover the phylogenetic
pathways that led from the Mesozoic biota, with its gymnosperm plants,
dinosaurs, opposite-birds, primitive mammals and so on, to the Cænozoic
arrays of lizards, snakes, modern birds, mammals and such? This
is a question of knowability. Or we might ask:
given a knowledge of the changing conditions on the earth during the Mesozoic
and Cænozoic, and of the characteristics of the Mesozoic biota, could
we deduce the demise of creatures like the dinosaurs, opposite-birds
and all, and their replacement by the Cænozoic array of forms?
This is a question of predictability.
These reflections recall a distinction alluded to earlier: that
of phylogenetic lineages from the characters which typify them at any point
in time. Lineages are, by their very nature, unique contingent entities,
defined by an uninterrupted information flow (inheritance), localised in
space and time, and possessed of particular arrays of characters which,
in ensemble, typify their form, at least over some span of time.
However, it is not at all clear whether, or to what extent, these characters
in themselves are unique, either in space or in time - as we
have seen, this depends greatly on how these characters are constructed,
and on their necessary connection with the unique genetic recipes that
generate them and define their lineage. It
is certainly evident that the characters of a long-lived lineage do not
define that lineage: they only typify it for some limited span of
time: we are of a lineage that includes what many would call fish,
and much else besides!
To illustrate this distinction, let us ask if, supposing those primitive
Mesozoic mammalian lineages had all been extinguished along with the dinosaurs
etc., would this mean that organisms with a warm-blooded physiology, with
hair, live-bearing, lactation, heterodont dentition, and behavioural complexity
- organisms that we recognise as modern mammals - could never have arisen
on earth? Ever? Or could that collection of characteristics
have arisen from some other, non-extinguished, reptilian lineage?
After all, it has ben seriously suggested that some of the dinosaurs and
related lineages may have developed homeothermy and hair, and heterodonty
has developed to varying degrees. If such evolution were
a possibility, then it would mean that, in this sense, "mammals", considered
simply as tetrapods with the above-mentioned characteristics, would not
be unique, contingent entities (and neither would any other lineage be,
defined simply in terms of such morphological characters - just as with
the Antillean lizards). It would also mean that large features of
evolution CAN repeat itself, contrary to widespread current opinion.
To drive this point home, would the extinction of all the Mesozoic mammals
have meant that a bipedal, behaviourally & socially complex organism,
with all the above characteristics and with the capacity for symbolic communication
& culture have been precluded, a case of "you simply can't get there
from here?" In short, if some creatures which we acknowledge
as already mammals had not survived the Mesozoic, would there be no creatures
on this planet discussing matters like the reconstruction of the past and
the meaning of life? Certainly the very lineage which
currently engages in such activities would not be here, for lineages are
indeed historically unique and contingent entities; but what of some
other lineage? after all, those we are happy to call mammals, primates,
hominids and such all do go back to some "reptile" lineage
- but just a different one from those which dominated the Mesozoic (one,
in fact, which dominated the late Paleozoic - the therapsids -who were
largely eclipsed at the end of that Era.)
The conclusions from this discussion may be formulated as follows:
Lineages are defined by an unbroken sequence of contingent primary texts
which permit their history to be knowable, yet neither their form nor their
fates are predictable therefrom; characters are not defined by any
single primary text, and so provide only ambiguous clues to history;
yet their fates are, in principle, predictable to the extent that they
impact on reproductive fitness.
A recently published proceedings (18)
of a symposium held in 1995 at McGill University, Canada, provides ample
evidence that the examples described here are in no way unusual.
The volume discusses many cases showing the same kinds of phenomena:
rampant convergence in a great diversity of organisms, from bats to monkeys,
from plants to fish, and more. As a closing thought on the relationship
between characters, ancestry and adaptation, I quote some words of Givnish
from his opening chapter in that volume which admirably render in concrete
terms the abstract conclusions of the previous paragraph: "We cannot
predict which lineage of herbaceous plants invading an oceanic island will
evolve the treelike habit; we can predict, however, that arborescent
plants will evolve from some lineage on that island, provided that
the climate supports productive vegetation."
NOTE ADDED 24 March 1999.
Recent
reports on experimental evolution with bacteria strongly endorses the
arguments made here that different genetic histories provide little impediment
to the evolution of near-identical phenotypic form. As Travisano
says: "History does have an effect," he says, "but not as much as
you might think it would." for the original papers see:
"Adaptive radiation in a heterogeneous environment" by Paul Rainey and
Michael Travisano, Nature, vol 394, p 69 (1998)
"Dynamics of adaptation and diversification" by Richard Lenski and Michael
Travisano, Proceedings of the National Academy of Science, vol 91, p 6808