This application utilizes relational databases that are implemented using Microsoft .NET technology.
The software system can be set up to work on one computer or in a network with multiple users.
Users initially activate the databases they desire to employ and provide a user specific name to individualize each database.
Pre-constructed database templates are provided which contain standardized data fields. Each database contains two additional user defined fields for custom data, if necessary.
As new samples are added to the inventory, each is provided with a unique laboratory identifier. This identifier is assigned automatically and sequentially.
In this way, all incoming reagents are appointed a laboratory name alias.
The systematic assignment of an alias allows rapid retrieval when a given reagent is required.
LINA also contains a number of useful search tools:
One general search allows database searches using up to four user defined criteria linked by the Boolean operators AND/OR/NOT.
A second more elaborate DNA search tool is incorporated into the oligonucleotide database.
This DNA search tool allows the user to input a DNA sequence and search for primers present in the database that would anneal to the target sequence.
In summary, LINA represents an easily implemented and useful organizational tool for biological laboratories with large numbers of strains, clones, or other reagents.
LINA was published in the Journal of Laboratory Automation.
A direct link to the publication is Yousef et al., JALA Volume 16, Issue 1, Pages 82-89, February 2011.

Screen shot of all four databases in LINA (ABOVE).
The four separate databases can be opened simultaneously and manipulated as individual windows.
They can be tiled vertically, horizontally, and cascaded using the “Windows” pull-down menu.

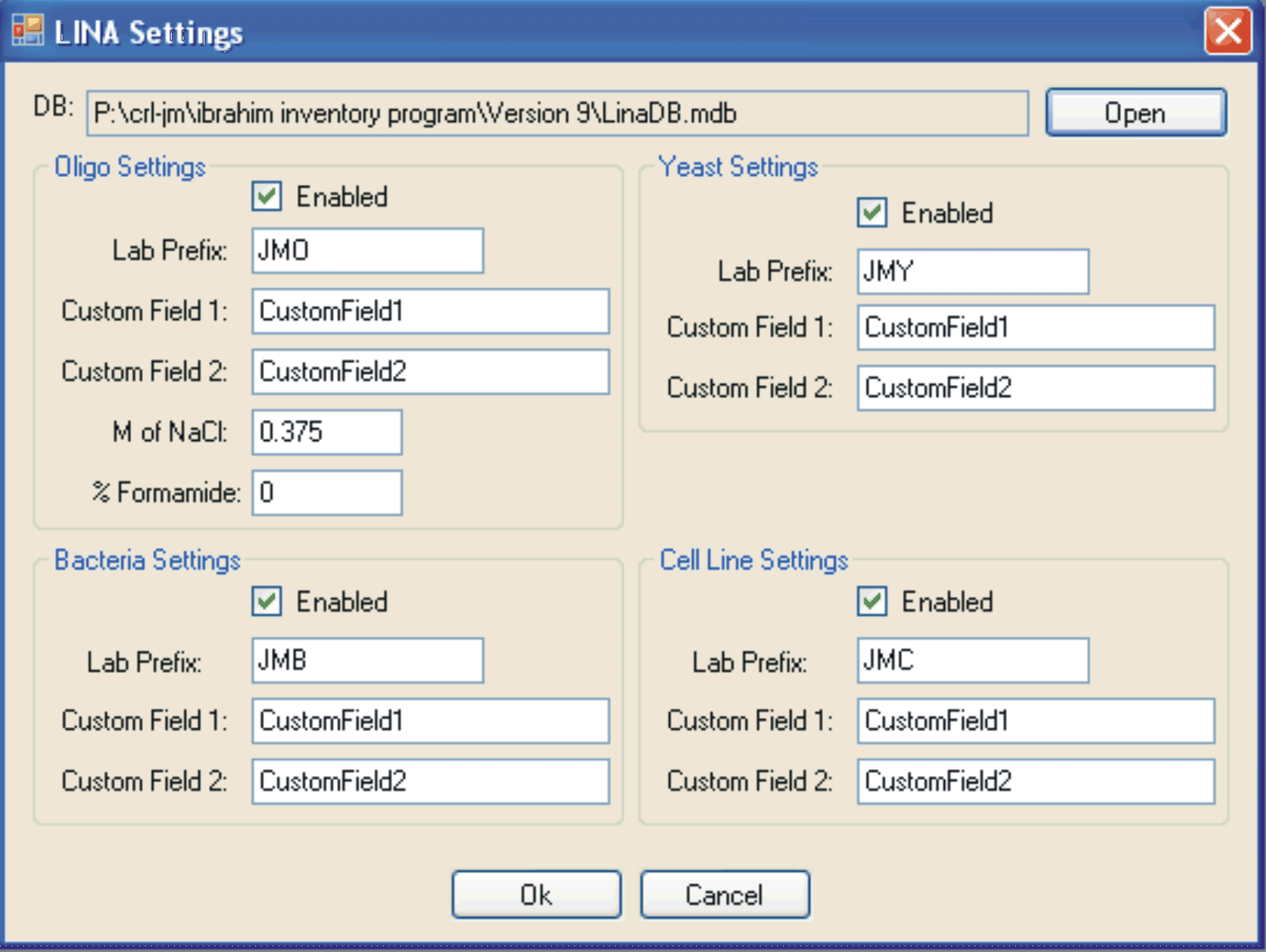
Screen shot of the settings menu for LINA (ABOVE).
Each individual database can be enabled, and the up to two custom fields can be added to each database.
Furthermore, specific NaCl and formamide concentrations can be entered in order to calculate the melting temperatures of the oligonucleotides in the “Oligo database”.
Finally, the location of the database file (*.mdb) is also determined in this menu which allows for different machines to point to a similar file thus enabling networking.
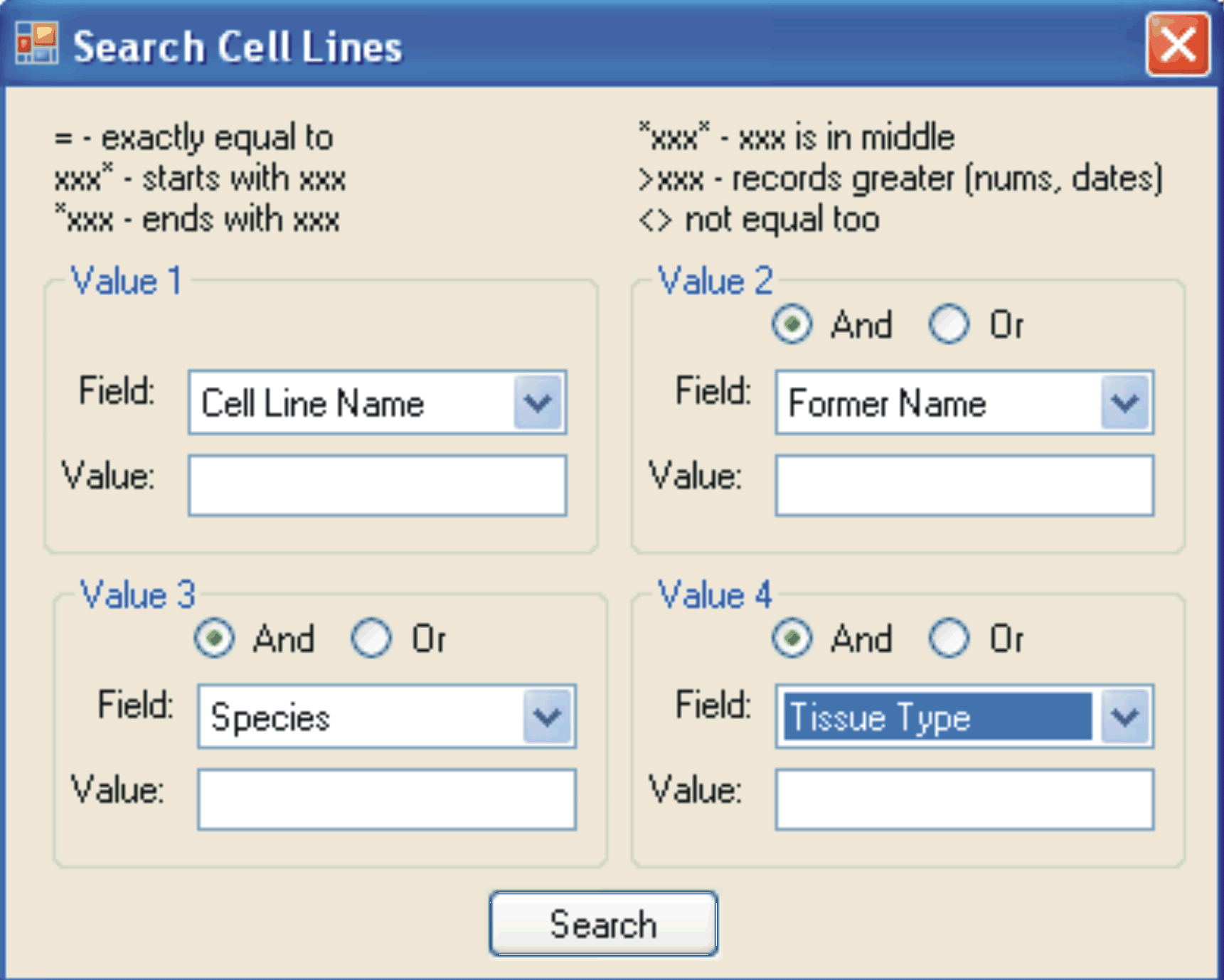
Screenshot of the search tool available for each database in LINA (ABOVE).
Up to four independent fields can be searched simultaneously using the AND/OR Boolean operators.
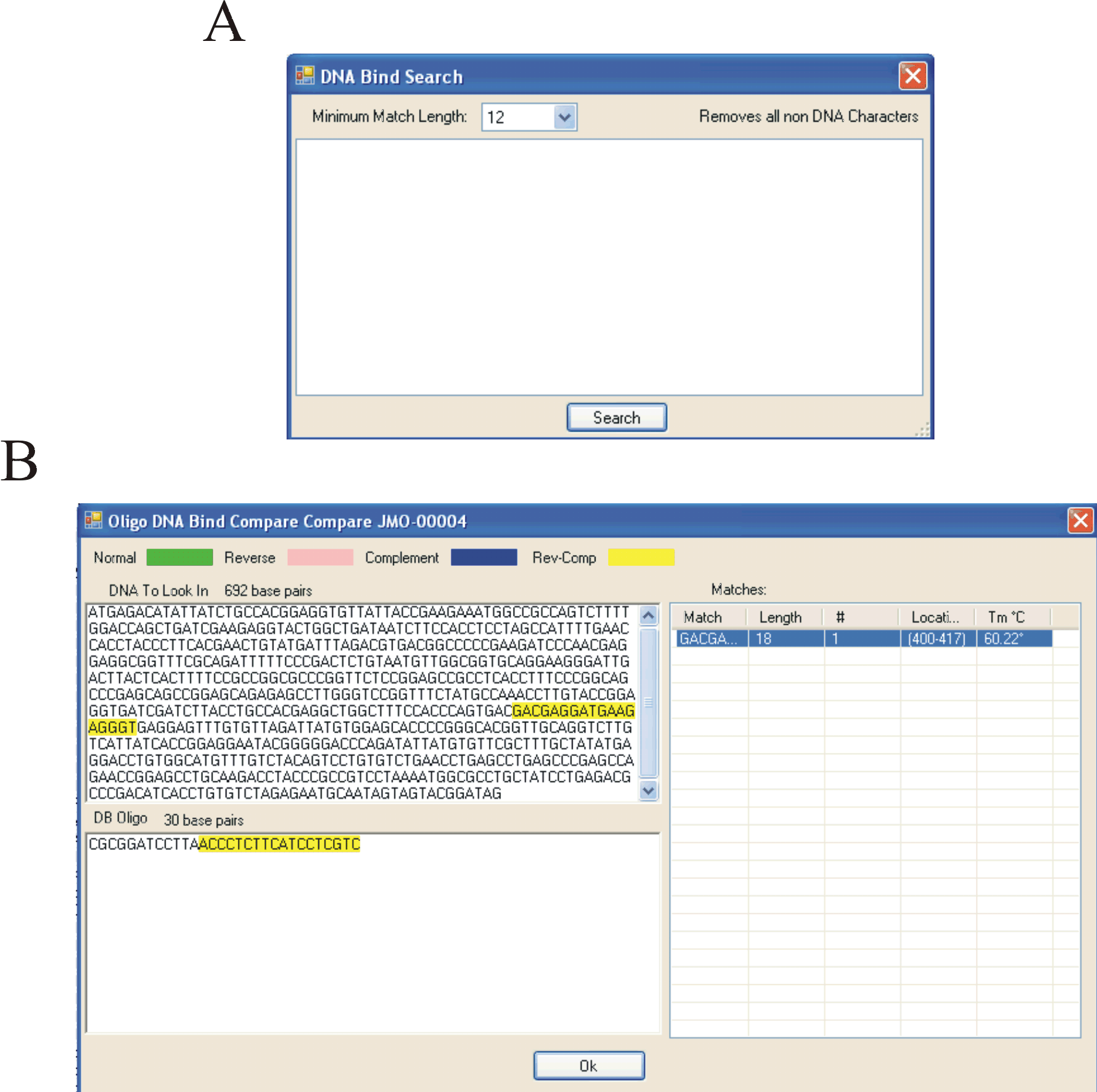
Screenshots of the Oligonucleotide database search tools (ABOVE).
A) The oligonucleotide database contains an additional “DNA Bind Search” which allows the user to search for identical, complimentary, reverse, or reverse complimentary matches to a given sequence.
B) The “DNA compare” function allows the user to see exactly where in the entered sequence an oligonucleotide binds.
In this screen shot, the Oligonuclotide JMO-0004 binds in a reverse complimentary way denoted by yellow to the entered sequence.
REMEMBER TO BACKUP YOUR DATABASE FREQUENTLY!