Keap1 plays a critical regulatory role in the cellular responses to oxidative stress through the interactions with multiple intrinsically disordered proteins, including Nrf2 and Prothymosin-alpha, via its Kelch domain. Failure in this defense mechanism can lead to pathological conditions such as cancer, cardiovascular disease, and neurodegenerative disorder. Through biophysical and biochemical analyses, we have extensively characterized the structure and dynamics of the intrinsically disordered Prothymosin-alpha (Yi et al. (2007) Biochemistry 46, 13120; F1000Prime recommended).
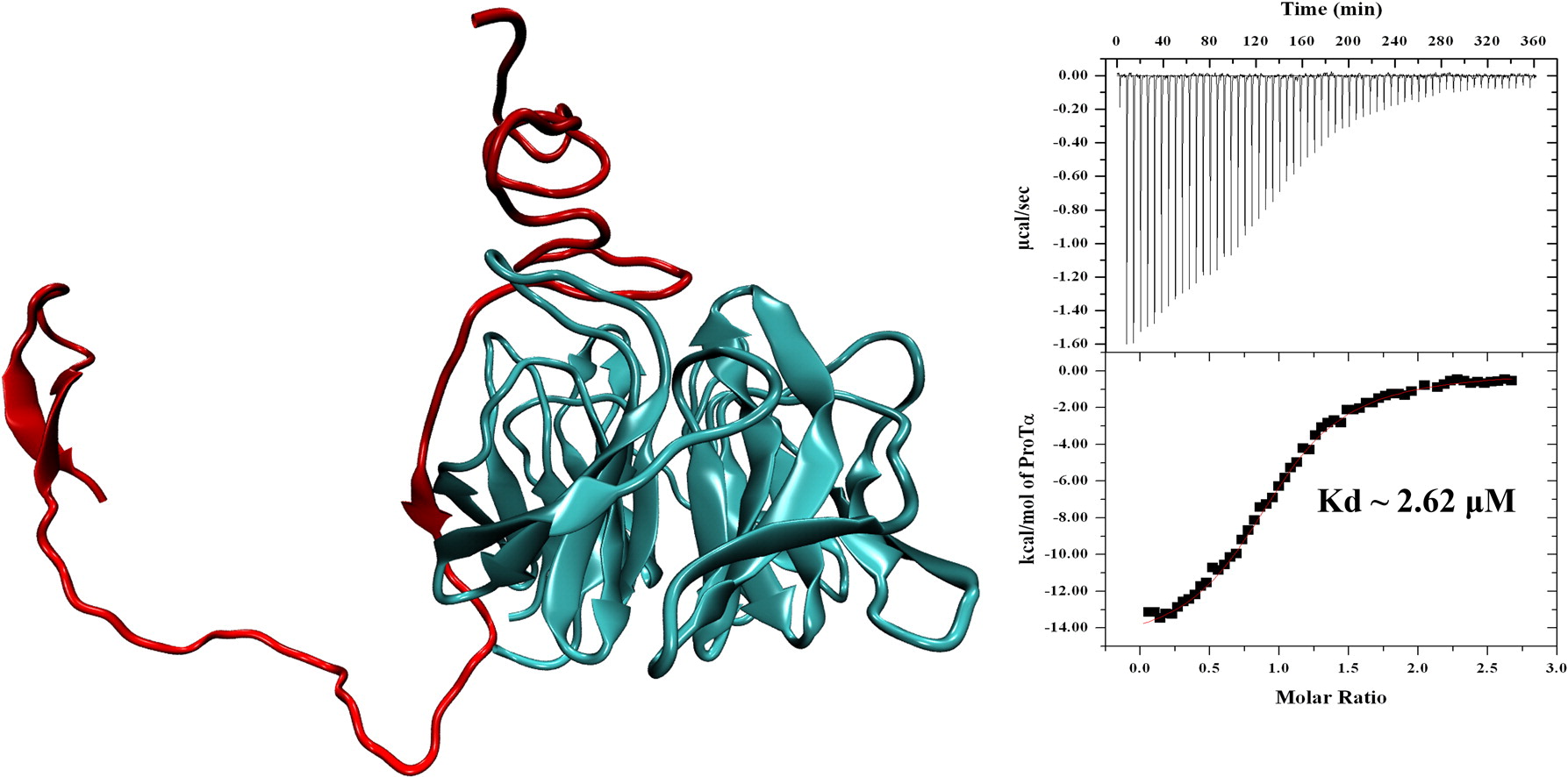
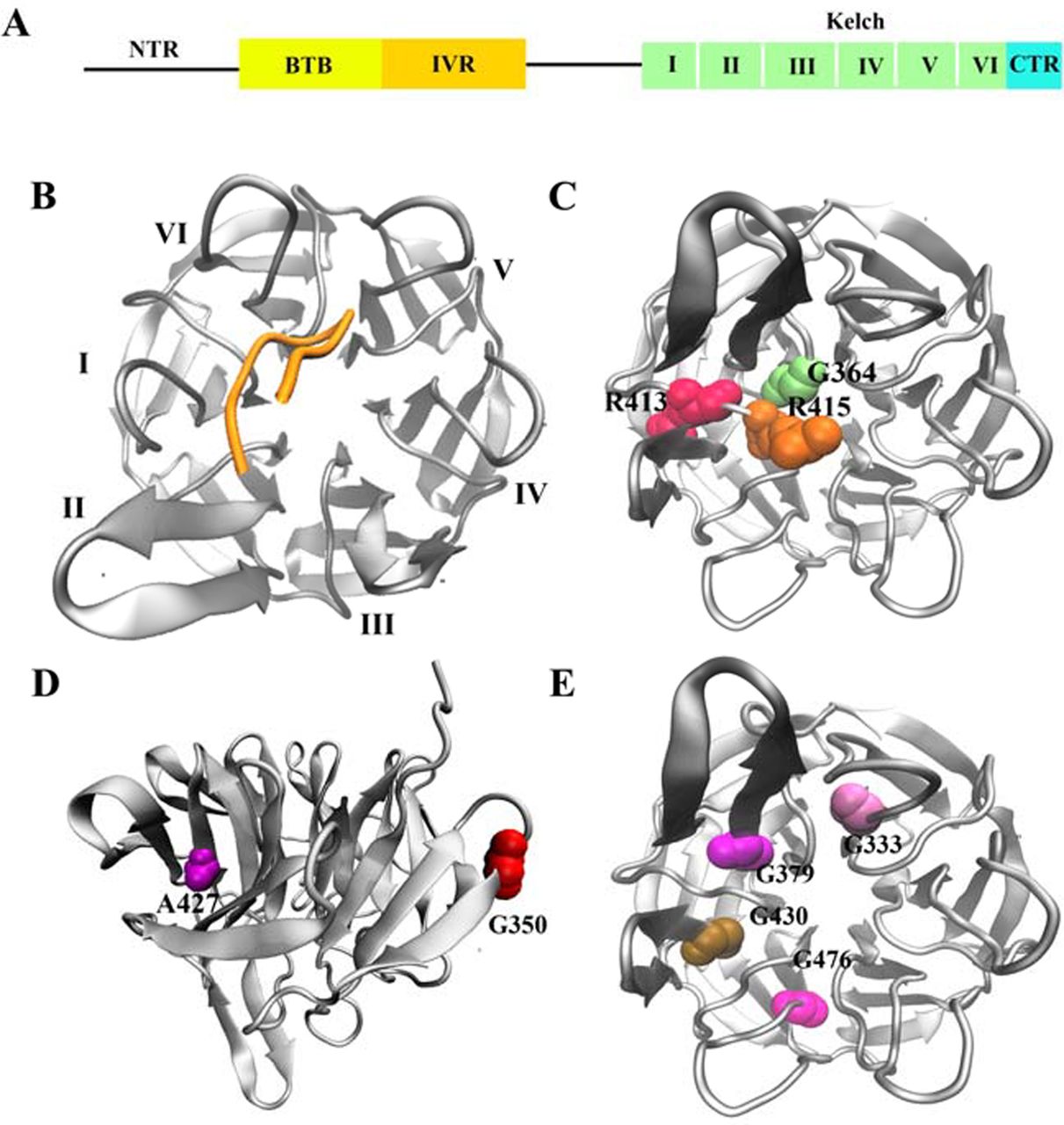
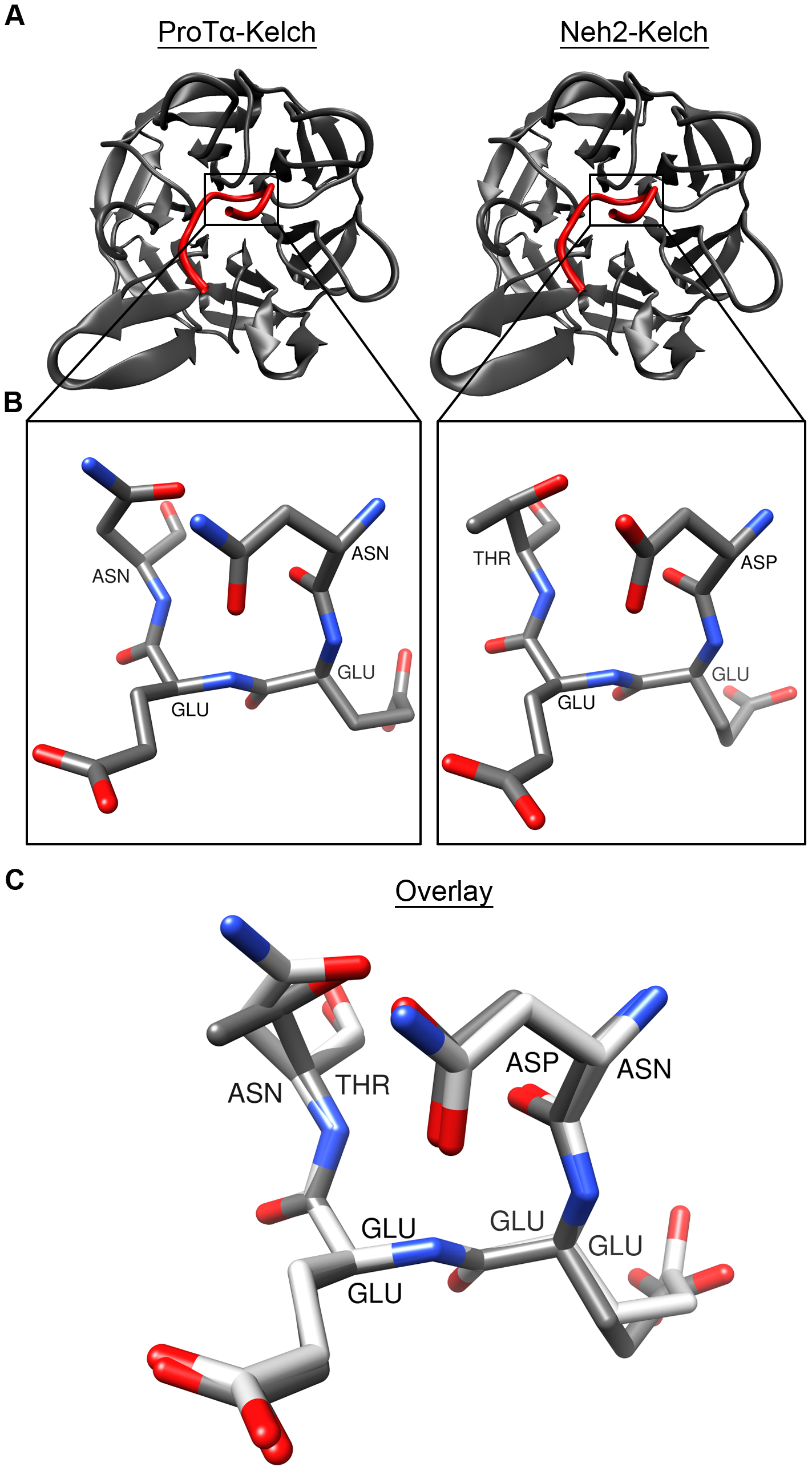
We also used a combination of experimental and computational approaches to dissect the mechanism by which Prothymosin-alpha interacts with Keap1 in oxidative stress response. We demonstrated that disordered Prothymosin binds to the Kelch domain of Keap1 in a fuzzy-complex manner. The result of this work, along with a few very recently characterized fuzzy complexes, calls for a new view of protein-protein interaction (Khan et al. (2013) Journal of Molecular Biology, 425, 1011; F1000Prime recommended). In recent work, we have characterized the molecular effects of a group of cancer somatic mutations of Keap1 on Nrf2-binding (Khan et al. (2015) Biochem J, 467, 141).
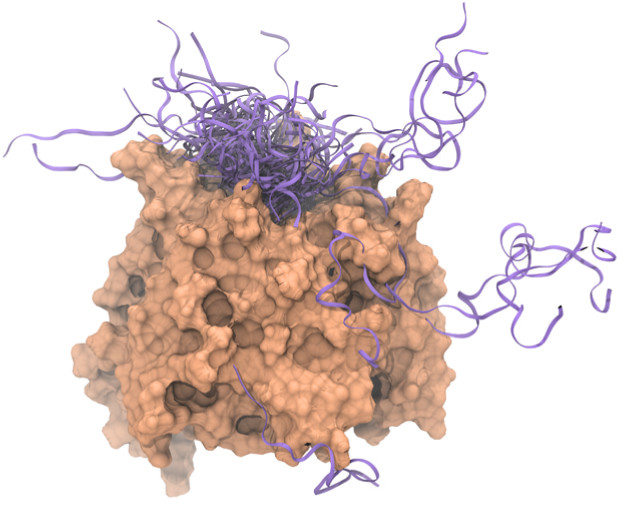
In collaboration with Dr. Mikko Karttunen (Eindhoven University of Technology, the Netherlands) we combine experimental and computational approaches to investigate the correlations of conformational propensity and dynamics of proteins to their specificities and affinities for targets (Cino et al. (2011) PLoS One, 6:e27371; Cino et al. (2013) Scientific Rep., 3:2305; Do et al. (2016) J Chem Theory Comput, 12, 395). Using this approach, we revealed the mechanisms by which Nrf2 binds to Keap1 in atomic detail. Furthermore, to facilitate the studies of intrinsically disordered proteins (IDPs) by MD simulations, we have extensively evaluated 10 different biomolecular force fields for the MD simulation of IDPs (Cino et al. (2012) J Chem Theory Comput, 8, 2725-2740). Such a comprehensive comparison offers excellent guidance to others conducting similar types of simulations. The paper is one of the journal’s Top 20 most downloaded articles during the first 12 months after its publication.
Our research on the structure-function relationship of Chibby in Wnt/beta-catenin signaling unveiled that the protein is partially disordered (Mokhtarzada et al. (2011) Biochemistry, 50, 715-726). This work provides the first structural and dynamic details of this novel antagonist in Wnt/-catenin signaling. We then scrutinized the molecular basis of the 14-3-3/Chibby interaction, a crucial complex in regulating Wnt signaling. Our study provides insight into how these two distinct molecular recognition features could facilitate disordered proteins in assembling complexes via different modes of interaction.
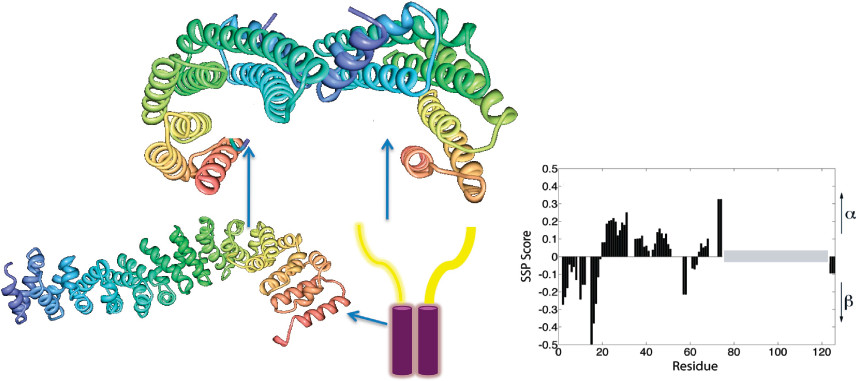
STI1P is well-known for its co-chaperone function with Hsp90. Recent studies show that this protein may also play a role in the pathogenesis of Alzheimer’s disease. In collaboration with Dr. Marco Prado (Robarts Research Institute, London, Canada), we use an interdisciplinary approach to dissect the mechanisms by which STI1P interacts with its protein targets in order to gain insight into its function.