|
|
|
Our laboratory explores the folding, structure, function and dynamics of proteins. Most of our work is based on the use of electrospray mass spectrometry (ESI-MS). Researchers working in our laboratory also have access to a wide range of other biophysical techniques, including high-field NMR, fluorescence, circular dichroism, UV-VIS, and stopped-flow spectroscopy, differential scanning calorimetry (DSC), analytical ultracentrifugation, surface-plasmon resonance (Biacore), and many others. A nanofabrication facility for the development of microfluidic devices is also available. Current research in our group includes the following topics:
Protein Folding Kinetics and Mechanisms The native state of a protein in solution is characterized by a tightly folded and highly specific three-dimensional structure. This native conformation is determined by the linear sequence of amino acids along the polypeptide backbone. In high concentrations of denaturants such as acid, proteins adopt a largely disordered (unfolded) conformation. Unfolding is reversible, i.e., the protein spontaneously refolds into its native structure when the denaturant is removed. This process occurs on the time scale of milliseconds to minutes. Protein folding is a remarkable phenomenon; there are billions of different possible ways to fold a protein chain and yet every protein "knows" how to find its native structure. Sometimes short-lived intermediates become populated during folding. The exact role of these intermediates for the overall folding mechanism is still a matter of debate. However, it is clear that the identification and structural characterization of these species is an important step towards a better understanding of protein folding. Electrospray ionization mass spectrometry (ESI-MS) is
one of the main techniques that is used in our lab. ESI generates
multiply protonated protein ions in the gas phase. "Time-resolved"
ESI-MS is a technique that allows the
folding kinetics and mechanisms to be studied in great detail.
Time-resolved ESI-MS can be used in conjunction with hydrogen/deuterium
exchange (HDX), thus
allowing the detailed characterization of folding intermediates. We
pursue both top-down and bottom-up MS approaches. Complementary
information can be obtained by laser-induced
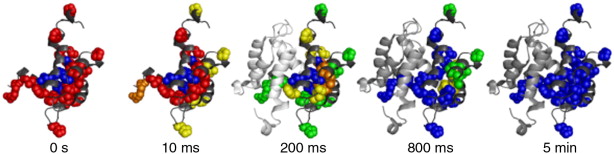
oxidative labeling. 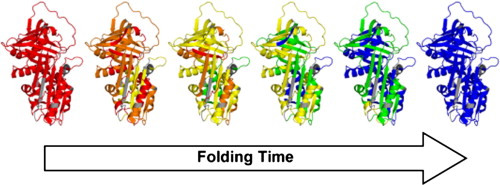 Figure
1: Time-resolved ESI-MS experiments provide "molecular
movies" that reveal how a protein folds. In this OH radical labeling
data set unfolded regions are shown in red, native elements are
depicted in blue.
Figure
2. In this experiment we used millisecond time-resolved
measurements to decipher the mechanism of a coupled folding/binding
reaction. The side chain solvent exposure ranges from high (red) to
native-like (blue). Protein Conformational Dynamics Proteins are highly dynamic systems that undergo
conformational fluctuations and unfolding/refolding transitions on
various time scales. In many cases, these conformational dynamics are
crucial for the biological function of a protein. Time-resolved
ESI-MS used in conjunction with HDX is a powerful tool for
monitoring these dynamics (Figure 3). We are studying the
behavior of proteins under conditions that lead to amyloid and other
forms of aggregates. This work has direct implications for
understanding the disease mechanisms of Alzheimer's and a
number of other disorders. The combination of top-down electron capture dissociation
(ECD) with HDX has opened up
new avenues for studying protein folding and dynamics.
Noncovalent Ligand-Protein Interactions Drug discovery and development relies on the identification of high-affinity ligands to specific target proteins. Our group has developed a novel ESI-MS-based approach for this purpose. Potential ligands are usually relatively small compounds that undergo rapid translational diffusion as long as they are free in solution. However, diffusion is drastically slowed down for ligands that are bound to a protein (Figure 4). We have devised a way of measuring the diffusion coefficients of a large number of potential ligands simultaneously by ESI-MS. It appears that this method could become an important tool for the high throughput screening of compound libraries. This approach is of significant interest to the pharmaceutical industry.
Development of Rapid-Mixing Systems for Kinetic Studies on Biochemical Systems ESI-MS has enormous potential for kinetic studies on a wide range of biochemical systems. Our laboratory is one of the few places worldwide where on-line MS-based kinetic studies can be carried out in the millisecond regime. We have developed a number of novel approaches in this area (Figure 5), and we are currently exploring the use of microfluidic systems to improve the time resolution of these techniques down to the sub-millisecond time range.
Enzyme Kinetics and Mechanisms Immediately after initiating an enzymatic reaction there is a
short time range (usually some milliseconds to seconds, depending on
the rate constants involved) during which the system approaches
steady-state conditions. The mechanism of the reaction dictates the
order by which short-lived intermediates become populated
successively during this period. Individual rate constants can be
measured from which the energy barriers along the reaction coordinate
can be estimated. Due to its high selectivity and sensitivity, time-resolved
ESI MS is a powerful tool for kinetic studies in this pre-steady-state
regime. In addition, we are interested in exploring the role of
enzyme conformational fluctuations
for the mechanism of catalysis.
Fundamental Aspects of Electrospray Ionization Despite the widespread use of ESI-MS, the exact nature of the
processes leading to the formation of gas phase ions from molecules in
solution is still not well understood. We are in the process of
deciphering
the physical reasons underlying the relationship between protein
conformations and ESI charge state distributions. Molecular dynamics (MD) simulations
turned out to be a key tool for gaining insights into the ESI
mechanism. Electrochemical
processes occurring during ESI represent another focus of our work.
 Figure 6. This frame is part of an MD trajectory
that shows how an ion is ejected from a highly charged ESI droplet.
Home | Lars Konermann | Research | People | Publications | News
|
||||||||||