
Mechanisms of Protein Ubiquitination in Health and Parkinson's Disease
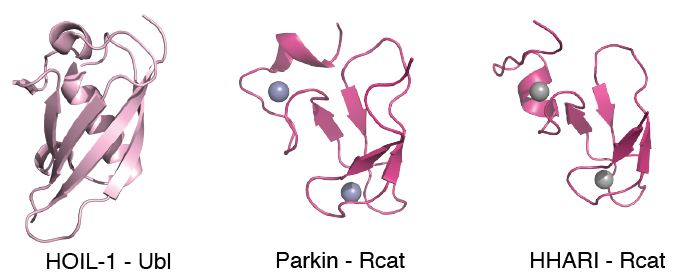
Genetic forms of Parkinson's disease (PD) are characterized by the loss of motor skills similar to the sporadic disease, a result of the death of dopaminergic neurons. One causative gene PARK2, encoding the RBR E3 ligase parkin, carries missense mutations that account for ~80% of early-onset PD. In conjunction with the E2 conjugating enzyme UbcH7, parkin labels unwanted proteins with ubiquitin (Ub) signalling the removal of damaged mitochondria, a proposed cause of PD. Parkin, and other RBR E3 ligases such as HHARI and HOIL-1 form a complicated set of multi-domain enzymes that must be stimulated to obtain maximal ligase activity. Recently, we determined the three-dimensional structures of several domains from these E3 ligase enzymes and showed that the Rcat domains from parkin and HHARI are necessary regions for ubiquitin catalysis.
Current projects in the lab focus on parkin and several other E2 conjugating and E3 ligase enzymes. In particular, we utilize biophysical methods to identify the interactions of E2 and E3 enzymes to understand the mechanisms these proteins use to function in the ubiquitination pathway.
Mechanisms of Membrane Repair and Trafficking by S100 Proteins
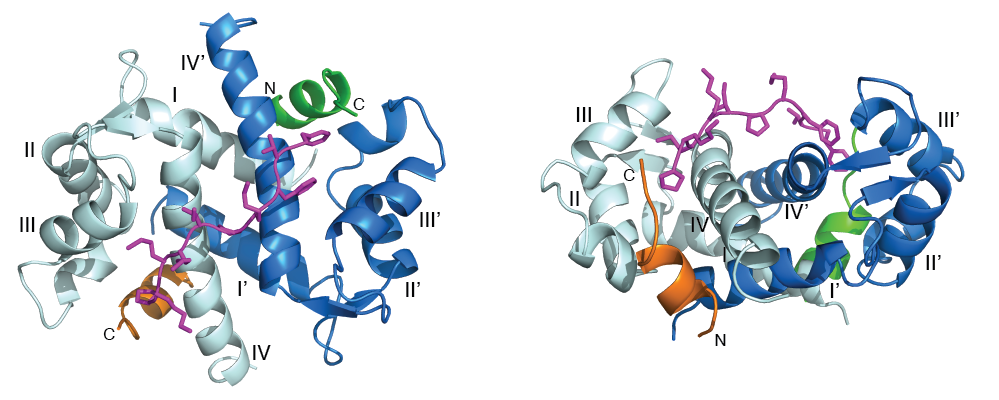
Calcium-sensor proteins are central regulators of human functions such as muscle contraction, neurotransmitter release and enzyme activation. One of the best characterized calcium-sensors is S100A10, a protein that forms a stable heterotetrameric complex with the phospholipid-binding protein annexin A2 (termed "A2t") and, along with the accessory proteins AHNAK and dysferlin, helps to repair ruptured plasma membranes in damaged cardiac and skeletal muscle. Dysfunction of proteins in this complex results in multiple forms of muscular dystrophy. A2t also preserves mood behavior balance by recruiting and translocating the serotonin 5-HT4 receptor to the plasma membrane. Disruption of this pathway causes neurological disorders such as depression. Recently, we engineered, and determined the structure of a heterotetrameric S100A10-annexin A2 chimeric protein in complex with a fragment from the membrane repair protein AHNAK as a first step to uncovering the mechanism of assembly of this important repair complex.
Current projects in the lab will use physical biochemical techniques such as high resolution NMR spectroscopy, fluorescence and isothermal calorimetry to explore how A2t and accessory proteins assemble the higher order dysferlin complex enabling muscle cell membrane repair. We also plan to use the S100A10 and A2t three-dimensional structures to rationally design small molecules that regulate A2t function.