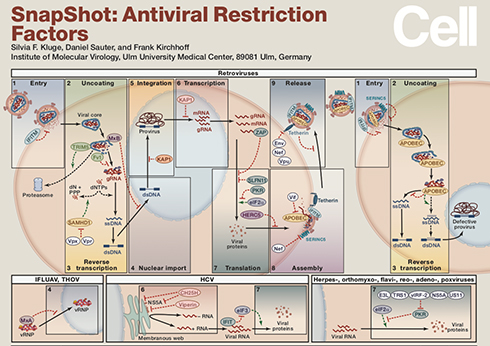
Publications
Featured publication
Communications Medicine volume 5, Article number: 405 (2025)
Early pandemic HIV-1 integration site preferences differ across anatomical sites
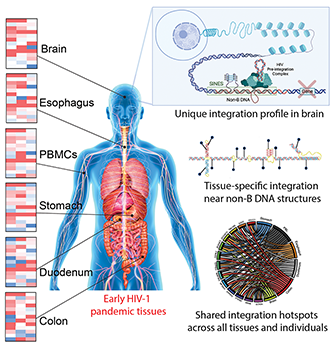 Abstract
Abstract
Pre-prints
None
Selected Publications
Click here for a full list of Dr. Barr's publications
(* corresponding author)
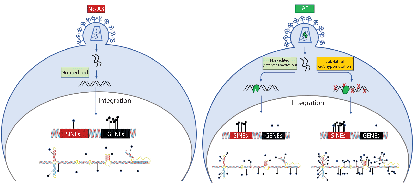
Nature Communications volume 14, Article number: 16 (2023)
Antiretroviral APOBEC3 Cytidine Deaminases Alter HIV-1 Provirus Integration Site Profiles
2Department of Biochemistry, Microbiology and Immunology, Faculty of Medicine, University of Ottawa, Ontario, Canada.
3uOttawa Center for Infection, Immunity and Inflammation (CI3).
†These authors contributed equally to this work.
 Abstract
Abstract
Biomolecules 2023, 13(7), 1095; https://doi.org/10.3390/biom13071095
Altmetrics
Editor's Choice
The Metallodrug BOLD-100 Is a Potent Inhibitor of SARS-CoV-2 Replication and Has Broad-Acting Antiviral Activity
Daniel Labach, Hinissan Kohio, Edwin Tse, Ermela Paparisto, Nicole Friesen, Jim Pankovich, Mark Bazett, and Stephen Barr*.
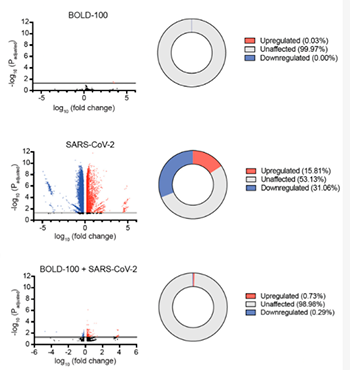
Abstract
The COVID-19 pandemic has highlighted an urgent need to discover and test new drugs to treat patients. Metal-based drugs are known to interact with DNA and/or a variety of proteins such as enzymes and transcription factors, some of which have been shown to exhibit anticancer and antimicrobial effects. BOLD-100 (sodium trans-[tetrachlorobis(1H-indazole)ruthenate(III)] dihydrate) is a novel ruthenium-based drug currently being evaluated in a Phase 1b/2a clinical trial for the treatment of advanced gastrointestinal cancer. Given that metal-based drugs are known to exhibit antimicrobial activities, we asked if BOLD-100 exhibits antiviral activity towards SARS-CoV-2. We demonstrated that BOLD-100 potently inhibits SARS-CoV-2 replication and cytopathic effects in vitro. An RNA sequencing analysis showed that BOLD-100 inhibits virus-induced transcriptional changes in infected cells. In addition, we showed that the antiviral activity of BOLD-100 is not specific for SARS-CoV-2, but also inhibits the replication of the evolutionarily divergent viruses Human Immunodeficiency Virus type 1 and Human Adenovirus type 5. This study identifies BOLD-100 as a potentially novel broad-acting antiviral drug.
Altmetrics
Targeting the MR1-MAIT cell axis improves vaccine efficacy and affords protection against viral pathogens.
1Department of Microbiology and Immunology, Western University, London, Ontario, Canada.
2McMaster Immunology Research Centre, McMaster University, Hamilton, Ontario, Canada.
3Krembil Centre for Stem Cell Biology, Molecular Medicine Research Laboratories, Robarts Research Institute, London, Ontario, Canada.
4Department of Physiology and Pharmacology, Western University, London, Ontario, Canada.
5Department of Medicine, McMaster University, Hamilton, Ontario, Canada.
6Department of Biochemistry and Biomedical Sciences, McMaster University, Hamilton, Ontario, Canada.
7Michael G. DeGroote Institute for Infectious Disease Research, McMaster University, Hamilton, Ontario, Canada.
8Division of Clinical Immunology and Allergy, Department of Medicine, Western University, London, Ontario, Canada.
9Division of General Surgery, Department of Surgery, Western University, London, Ontario, Canada.
10Lawson Health Research Institute, London, Ontario, Canada.
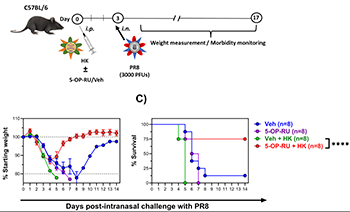 Abstract
Abstract
Mucosa-associated invariant T (MAIT) cells are MR1-restricted, innate-like T lymphocytes with tremendous antibacterial and immunomodulatory functions. Additionally, MAIT cells sense and respond to viral infections in an MR1-independent fashion. However, whether they can be directly targeted in immunization strategies against viral pathogens is unclear. We addressed this question in multiple wild-type and genetically altered but clinically relevant mouse strains using several vaccine platforms against influenza viruses, poxviruses and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). We demonstrate that 5-(2-oxopropylideneamino)-6-D-ribitylaminouracil (5-OP-RU), a riboflavin-based MR1 ligand of bacterial origin, can synergize with viral vaccines to expand MAIT cells in multiple tissues, reprogram them towards a pro-inflammatory MAIT1 phenotype, license them to bolster virus-specific CD8+ T cell responses, and potentiate heterosubtypic anti-influenza protection. Repeated 5-OP-RU administration did not render MAIT cells anergic, thus allowing for its inclusion in prime-boost immunization protocols. Mechanistically, tissue MAIT cell accumulation was due to their robust proliferation, as opposed to altered migratory behavior, and required viral vaccine replication competency and Toll-like receptor 3 and type I interferon receptor signaling. The observed phenomenon was reproducible in female and male mice, and in both young and old animals. It could also be recapitulated in a human cell culture system in which peripheral blood mononuclear cells were exposed to replicating virions and 5-OP-RU. In conclusion, although viruses and virus-based vaccines are devoid of the riboflavin biosynthesis machinery that supplies MR1 ligands, targeting MR1 enhances the efficacy of vaccine-elicited antiviral immunity. We propose 5-OP-RU as a non-classic but potent and versatile vaccine adjuvant against respiratory viruses.
iScience 2023, 26(4):106292. doi: 10.1016/j.isci.2023.106292.
Altmetrics
Preprint Altmetric
Monovalent and trivalent VSV-based COVID-19 vaccines elicit neutralizing antibodies and CD8+ T cells against SARS-CoV-2 variants.
Kate A Parham, Gyoung Nyoun Kim, Connor G Richer, Marina Ninkov, Kunyu Wu, Nasrin Saeedian, Yue Li, Rasheduzzaman Rashu, Stephen D Barr, Eric J Arts, S M Mansour Haeryfar, C Yong Kang, Ryan M Troyer*.
Western University, Schulich School of Medicine and Dentistry, Department of Microbiology and Immunology, London, Ontario, Canada.
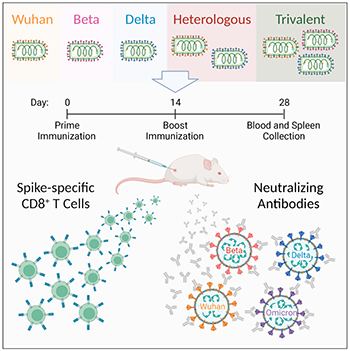 Abstract
Abstract
Recombinant vesicular stomatitis virus (rVSV) vaccines expressing spike proteins of Wuhan, Beta, and/or Delta variants of SARS-CoV-2 were generated and tested for induction of antibody and T cell immune responses following intramuscular delivery to mice. rVSV-Wuhan and rVSV-Delta vaccines and an rVSV-Trivalent (mixed rVSV-Wuhan, -Beta, -Delta) vaccine elicited potent neutralizing antibodies (nAbs) against live SARS-CoV-2 Wuhan (USAWA1), Beta (B.1.351), Delta (B.1.617.2), and Omicron (B.1.1.529) viruses. Prime-boost vaccination with rVSV-Beta was less effective in this capacity. Heterologous boosting of rVSV-Wuhan with rVSV-Delta induced strong nAb responses against Delta and Omicron viruses, with the rVSV-Trivalent vaccine consistently effective in inducing nAbs against all the SARS-CoV-2 variants tested. All vaccines, including rVSV-Beta, elicited a spike-specific immunodominant CD8+ T cell response. Collectively, rVSV vaccines targeting SARS-CoV-2 variants of concern may be considered in the global fight against COVID-19.
Viruses 2023, 15(2):465. https://doi.org/10.3390/v15020465
Altmetrics
Specialized DNA structures act as genomic beacons for integration by evolutionarily diverse retroviruses
Hinissan P Kohio†, Hannah O Ajoge†, Macon D Coleman, Emmanuel Ndashimye, Richard M Gibson, Eric J Arts, Stephen Dominic Barr*
Western University, Schulich School of Medicine and Dentistry, Department of Microbiology and Immunology, London, Ontario, Canada.
†These authors contributed equally to this work.
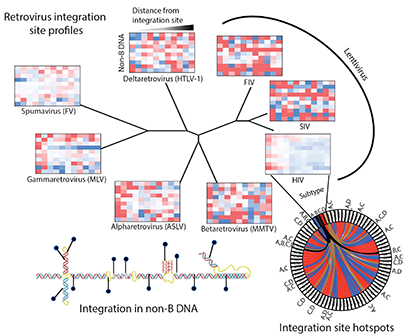 Abstract
Abstract
Retroviral integration site targeting is not random and plays a critical role in expression and long-term survival of the integrated provirus. To better understand the genomic environment surrounding retroviral integration sites, we performed a meta-analysis of previously published integration site data from evolutionarily diverse retroviruses, including new experimental data from HIV-1 subtypes A, B, C and D. We show here that evolutionarily divergent retroviruses exhibit distinct integration site profiles with strong preferences for integration near non-canonical B-form DNA (non-B DNA). We also show that in vivo-derived HIV-1 integration sites are significantly more enriched in transcriptionally silent regions and transcription-silencing non-B DNA features of the genome compared to in vitro-derived HIV-1 integration sites. Integration sites from individuals infected with HIV-1 subtype A, B, C or D viruses exhibited different preferences for common genomic and non-B DNA features. In addition, we identified several integration site hotspots shared between different HIV-1 subtypes, all of which were located in the non-B DNA feature slipped DNA. Together, these data show that although evolutionarily divergent retroviruses exhibit distinct integration site profiles, they all target non-B DNA for integration. These findings provide new insight into how retroviruses integrate into genomes for long-term survival.
Viruses. 2022 Nov 11;14(11):2494. doi: 10.3390/v14112494.
Altmetrics
G-Quadruplex DNA and Other Non-Canonical B-Form DNA Motifs Influence Productive and Latent HIV-1 Integration and Reactivation Potential
Hannah O Ajoge, Hinissan P Kohio, Ermela Paparisto, Macon D Coleman, Kemen Wong, Sean K Tom, Katie L Bain, Charles C Berry, Eric J Arts, Stephen D Barr*
Abstract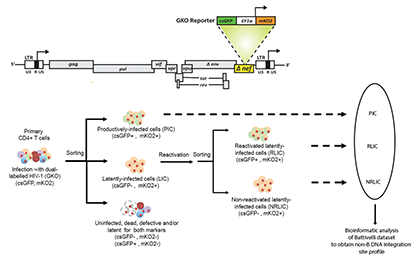 The integration of the HIV-1 genome into the host genome is an essential step in the life cycle of the virus and it plays a critical role in the expression, long-term persistence, and reactivation of HIV expression. To better understand the local genomic environment surrounding HIV-1 proviruses, we assessed the influence of non-canonical B-form DNA (non-B DNA) on the HIV-1 integration site selection. We showed that productively and latently infected cells exhibit different integration site biases towards non-B DNA motifs. We identified a correlation between the integration sites of the latent proviruses and non-B DNA features known to potently influence gene expression (e.g., cruciform, guanine-quadruplex (G4), triplex, and Z-DNA). The reactivation potential of latent proviruses with latency reversal agents also correlated with their proximity to specific non-B DNA motifs. The perturbation of G4 structures in vitro using G4 structure-destabilizing or -stabilizing ligands resulted in a significant reduction in integration within 100 base pairs of G4 motifs. The stabilization of G4 structures increased the integration within 300-500 base pairs from G4 motifs, increased integration near transcription start sites, and increased the proportion of latently infected cells. Moreover, we showed that host lens epithelium-derived growth factor (LEDGF)/p75 and cleavage and polyadenylation specificity factor 6 (CPSF6) influenced the distribution of integration sites near several non-B DNA motifs, especially G4 DNA. Our findings identify non-B DNA motifs as important factors that influence productive and latent HIV-1 integration and the reactivation potential of latent proviruses.
The integration of the HIV-1 genome into the host genome is an essential step in the life cycle of the virus and it plays a critical role in the expression, long-term persistence, and reactivation of HIV expression. To better understand the local genomic environment surrounding HIV-1 proviruses, we assessed the influence of non-canonical B-form DNA (non-B DNA) on the HIV-1 integration site selection. We showed that productively and latently infected cells exhibit different integration site biases towards non-B DNA motifs. We identified a correlation between the integration sites of the latent proviruses and non-B DNA features known to potently influence gene expression (e.g., cruciform, guanine-quadruplex (G4), triplex, and Z-DNA). The reactivation potential of latent proviruses with latency reversal agents also correlated with their proximity to specific non-B DNA motifs. The perturbation of G4 structures in vitro using G4 structure-destabilizing or -stabilizing ligands resulted in a significant reduction in integration within 100 base pairs of G4 motifs. The stabilization of G4 structures increased the integration within 300-500 base pairs from G4 motifs, increased integration near transcription start sites, and increased the proportion of latently infected cells. Moreover, we showed that host lens epithelium-derived growth factor (LEDGF)/p75 and cleavage and polyadenylation specificity factor 6 (CPSF6) influenced the distribution of integration sites near several non-B DNA motifs, especially G4 DNA. Our findings identify non-B DNA motifs as important factors that influence productive and latent HIV-1 integration and the reactivation potential of latent proviruses.
PLoS Pathog. 2021 Dec 16;17(12):e1010092. doi: 10.1371/journal.ppat.1010092
Altmetrics
A vesicular stomatitis virus-based prime-boost vaccination strategy induces potent and protective neutralizing antibodies against SARS-CoV-2
Gyoung Nyoun Kim, Jung-ah Choi, Kunyu Wu, Nasrin Saeedian, Eunji Yang, Hayan Park, Sun-Je Woo, Gippeum Lim, Seong-Gyu Kim, Su-Kyeong Eo, Hoe Won Jeong, Taewoo Kim, Jae-Hyung Chang, Sang Hwan Seo, Na Hyung Kim, Eunsil Choi, Seungho Choo, Sangkyun Lee, Andrew Winterborn, Yue Li, Kate Parham, Justin M. Donovan, Brock Fenton, Jimmy D. Dikeakos, Gregory A. Dekaban, S. M. Mansour Haeryfar, Ryan M. Troyer, Eric J. Arts, Stephen D. Barr, Manki Song, C. Yong Kang
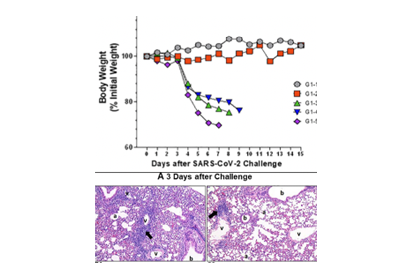 Summary
Summary
The development of safe and effective vaccines to prevent SARS-CoV-2 infections remains an urgent priority worldwide. We have used a recombinant vesicular stomatitis virus (rVSV)-based prime-boost immunization strategy to develop an effective COVID-19 vaccine candidate. We have constructed VSV genomes carrying exogenous genes resulting in the production of avirulent rVSV carrying the full-length spike protein (SF), the S1 subunit, or the receptor-binding domain (RBD) plus envelope (E) protein of SARS-CoV-2. Adding the honeybee melittin signal peptide (msp) to the N-terminus enhanced the protein expression, and adding the VSV G protein transmembrane domain and the cytoplasmic tail (Gtc) enhanced protein incorporation into pseudotype VSV. All rVSVs expressed three different forms of SARS-CoV-2 spike proteins, but chimeras with VSV-Gtc demonstrated the highest rVSV-associated expression. In immunized mice, rVSV with chimeric S protein-Gtc derivatives induced the highest level of potent neutralizing antibodies and T cell responses, and rVSV harboring the full-length msp-SF-Gtc proved to be the superior immunogen. More importantly, rVSV-msp-SF-Gtc vaccinated animals were completely protected from a subsequent SARS-CoV-2 challenge. Overall, we have developed an efficient strategy to induce a protective response in SARS-CoV-2 challenged immunized mice. Vaccination with our rVSV-based vector may be an effective solution in the global fight against COVID-19.
https://news.westernu.ca/2021/12/novel-covid-19-vaccine-shows-promise/
Altmetrics
Interferon-induced HERC5 Inhibits Ebola Virus Particle Production and Is Antagonized by Ebola Glycoprotein
Ermela Paparisto, Nina R Hunt, Daniel S Labach, Macon D Coleman, Eric J Di Gravio, Mackenzie J Dodge, Marceline Côté, Andreas Müller, Thomas Hoenen, Stephen D Barr *
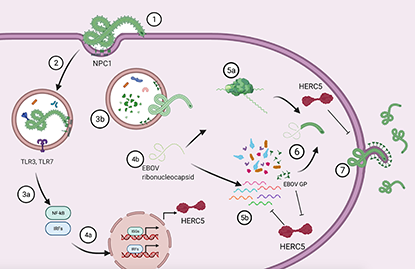 Summary
Summary
Survival following Ebola virus (EBOV) infection correlates with the ability to mount an early and robust interferon (IFN) response. The host IFN-induced proteins that contribute to controlling EBOV replication are not fully known. Among the top genes with the strongest early increases in expression after infection in vivo is IFN-induced HERC5. Using a transcription- and replication-competent VLP system, we showed that HERC5 inhibits EBOV virus-like particle (VLP) replication by depleting EBOV mRNAs. The HERC5 RCC1-like domain was necessary and sufficient for this inhibition and did not require zinc finger antiviral protein (ZAP). Moreover, we showed that EBOV (Zaire) glycoprotein (GP) but not Marburg virus GP antagonized HERC5 early during infection. Our data identifies a novel ‘protagonist-antagonistic’ relationship between HERC5 and GP in the early stages of EBOV infection that could be exploited for the development of novel antiviral therapeutics.
Journal of Antimicrobial Chemotherapy 2021 Aug 28;dkab276
High level resistance to bictegravir and cabotegravir in subtype A and D infected HIV-1 patients failing raltegravir with multiple primary resistance mutations.
Emmanuel Ndashimye, Yue Li, Paul S Reyes, Mariano Avino, Abayomi S Olabode, Cissy M Kityo, Fred Kyeyune, Immaculate Nankya, Miguel E Quiñones-Mateu, Stephen D Barr, Eric J Arts*
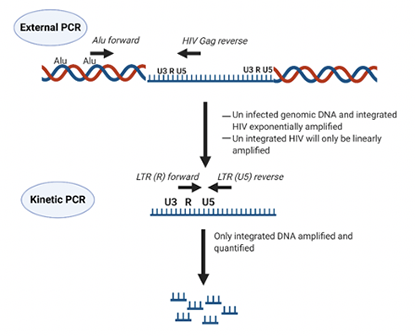
Summary
The second-generation integrase strand transfer inhibitor (INSTI) bictegravir (BIC) is becoming accessible in low- and middle-income countries (LMICs), and another INSTI (cabotegravir, CAB) is in late-stage clinical trials. Data on BIC and CAB susceptibility in raltegravir (RAL)-experienced HIV-1 non-B subtype infected patients carrying drug resistance mutations (DRMs) remains very scarce in LMICs. HIV-1 integrase (INT) recombinant viruses from eight patients failing RAL-based third-line therapy in Uganda were genotypically and phenotypically tested for susceptibility to BIC and CAB. Ability of these viruses to integrate into human genomes was assessed in MT-4 cells. HIV-1 INT-recombinant viruses harboring single primary mutations (N155H or Y143R/S) or in combination with secondary INSTI mutations, T97A, M50I, L74IM, E157Q, G163R, or V151I were susceptible to both BIC and CAB. However, combinations of primary INSTI-resistance mutations such as E138A/G140A/G163R/Q148R or E138K/G140A/S147G/Q148K led to decreased susceptibility to both CAB (fold change in EC50 values from 429 to 1,000x) and BIC (60 to 100x), exhibiting a high degree of cross resistance. Moreover, these same INT-recombinant viruses showed impaired integration capacity (14% to 48%) relative to the wild type HIV-1 NL4-3 strain in the absence of drug. Though not currently utilized in most LMICs, BIC offers a valid alternative to HIV-infected individuals harboring subtype A and D HIV-1 variants with reduced susceptibility to first generation INSTIs but previous exposure to RAL may reduce efficacy.
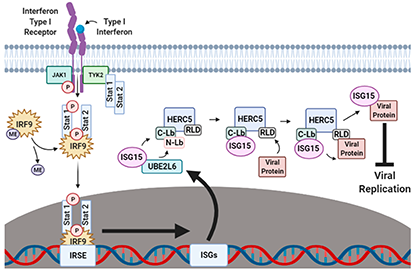
HERC5 and the ISGylation Pathway: Critical Modulators of the Antiviral Immune Response
Nicholas A. Mathieu, Ermela Paparisto, Stephen D. Barr and Donald E. Spratt*
 Summary
Summary
Mammalian cells have developed an elaborate network of immunoproteins that serve to identify and combat viral pathogens. Interferon-stimulated gene 15 (ISG15) is a 15.2 kDa tandem ubiquitin-like protein (UBL) that is used by specific E1–E2–E3 ubiquitin cascade enzymes to interfere with the activity of viral proteins. Recent biochemical studies have demonstrated how the E3 ligase HECT and RCC1-containing protein 5 (HERC5) regulates ISG15 signaling in response to hepatitis C (HCV), influenza-A (IAV), human immunodeficiency virus (HIV), SARS-CoV-2 and other viral infections. Taken together, the potent antiviral activity displayed by HERC5 and ISG15 make them promising drug targets for the development of novel antiviral therapeutics that can augment the host antiviral response. In this review, we examine the emerging role of ISG15 in antiviral immunity with a particular focus on how HERC5 orchestrates the specific and timely ISGylation of viral proteins in response to infection.
Research Outreach 2019 Feb; ISSN 2517-7028, Issue 106
Evolutionary arms race: A 400 million-year-old battle between HIV and ancient genes, HERC5 and HERC6
Ermela Paparisto and Stephen D. Barr* Summary
Summary
Two human genes, HERC5 and HERC6 have protective effects against HIV and other viruses. Dr Stephen D. Barr from Western University, Canada, traces the evolution of these genes throughout human history and investigates their protective effects. His work has shown that these genes are involved in an evolutionary arms race, with host defence mechanisms and the virus ever trying to outwit one another. Increased understanding of the viral mechanisms involved may present novel therapeutic targets for antiviral treatments.
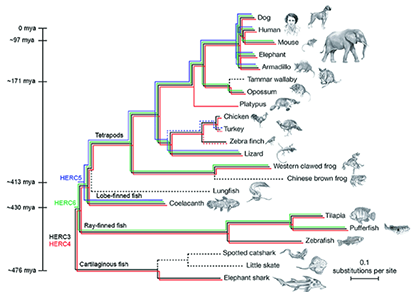
Evolution-guided structural and functional analyses of the HERC family reveals an ancient marine origin and determinants of antiviral activity
 Abstract
Abstract
In humans, 'homologous to the E6-AP carboxyl terminus (HECT) and regulator of chromosome condensation 1 (RCC1)-like domain-containing protein 5' (HERC5) is an interferon-induced protein that inhibits replication of evolutionarily diverse viruses including human immunodeficiency virus type 1 (HIV-1). To better understand the origin, evolution and function of HERC5, we performed phylogenetic, structural and functional analyses of the entire human small HERC family, which includes HERC3, HERC4, HERC5 and HERC6. We demonstrated that the HERC family emerged >595 million years ago and has undergone gene duplication and gene loss events throughout its evolution. The structural topology of the RCC1-like domain and HECT domains from all HERC paralogs are highly conserved among evolutionarily diverse vertebrates despite low sequence homology. Functional analyses showed that the human small HERCs exhibit different degrees of antiviral activity towards HIV-1 and that HERC5 provided the strongest level of inhibition. Notably, coelacanth HERC5 inhibited SIV, but not HIV-1, particle production suggesting that the antiviral activity of HERC5 emerged over 413 million years ago and exhibits species- and virus-specific restriction. In addition, we showed that both HERC5 and HERC6 are evolving under strong positive selection; particularly blade 1 of the RCC1-like domain, which we showed is a key determinant of antiviral activity. These studies provide insight into the origin, evolution and biological importance of the human restriction factor HERC5 and the other HERC family members.
IMPORTANCE Intrinsic immunity plays an important role as the first line of defense against viruses. Studying the origin, evolution and function of proteins responsible for effecting this defense will provide key information about virus-host relationships that can be exploited for future drug development. We showed that HERC5 is one such antiviral protein that belongs to an evolutionarily conserved family of HERCs with an ancient marine origin. Not all vertebrates possess all HERC members, suggesting that different HERCs emerged at different times during evolution to provide the host with a survival advantage. Consistent with this, some of the more recently emerged HERC members, HERC5 and HERC6, displayed strong signatures of having been involved in an ancient evolutionary battle with viruses. Our findings provide new insights into the evolutionary origin and function of the HERC family in vertebrate evolution, identifying HERC5 and possibly HERC6 as important effectors of intrinsic immunity in vertebrates.
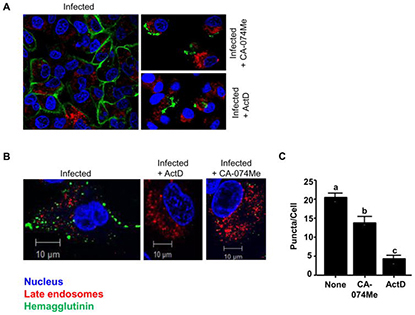
Cathepsin B plays a key role in optimal production of the influenza A virus
 Abstract
Abstract
Background: Influenza A- virus (IAV) is the etiologic agent of the febrile respiratory illness, commonly referred to as ‘flu’. The lysosomal protease cathepsin B (CTSB) has shown to be involved in the lifecycle of various viruses. Here, we examined the role of CTSB in the IAV lifecycle. Methods: CTSB-deficient (CTSB -/- ) macrophages and the human lung epithelial cell line A549 cells treated with CA-074Me were infected with the A/Puerto Rico/8/34 strain of IAV (IAV- PR8). Viral entry and propagation were measured through quantitative realtime RT-PCR; production and localization of hemagglutinin (HA) protein in the infected host cells were analysed by Western blots, flow cytometry and confocal microscopy; production of progeny viruses were measured by a hemagglutination assay. Results: CTSB -/- macrophages and CA-074Me-treated A549 cells had no defects in incorporating IAV-PR8 virions and permitting viral RNA synthesis. However, these cells produced significantly lower amounts of HA protein and progeny virions than wild-type or untreated cells. Conclusion: These data indicate that CTSB is involved in the expression of IAV-PR8 HA protein and subsequent optimal production of IAV-PR8 progeny virions. Targeting CTSB can be a novel therapeutic strategy for treating IAV infection.
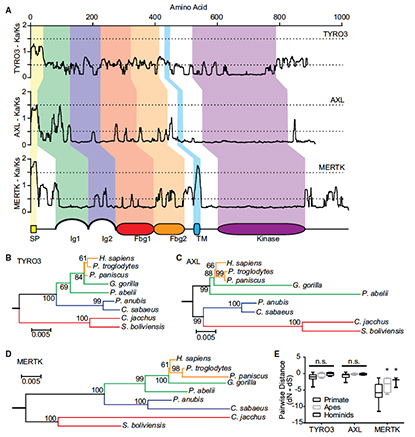
Antagonistic Coevolution of MER Tyrosine Kinase Expression and Function.
Abstract
Mol Ther Nucleic Acids. 2014 Aug 26;3:e187. doi: 10.1038/mtna.2014.39.Lentivector integration sites in ependymal cells from a model of metachromatic leukodystrophy: non-B DNA as a new factor influencing integration.
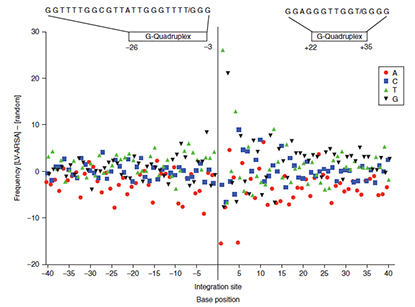
Abstract
The blood-brain barrier controls the passage of molecules from the blood into the central nervous system (CNS) and is a major challenge for treatment of neurological diseases. Metachromatic leukodystrophy is a neurodegenerative lysosomal storage disease caused by loss of arylsulfatase A (ARSA) activity. Gene therapy via intraventricular injection of a lentiviral vector is a potential approach to rapidly and permanently deliver therapeutic levels of ARSA to the CNS. We present the distribution of integration sites of a lentiviral vector encoding human ARSA (LV-ARSA) in murine brain choroid plexus and ependymal cells, administered via a single intracranial injection into the CNS. LV-ARSA did not exhibit a strong preference for integration in or near actively transcribed genes, but exhibited a strong preference for integration in or near satellite DNA. We identified several genomic hotspots for LV-ARSA integration and identified a consensus target site sequence characterized by two G-quadruplex-forming motifs flanking the integration site. In addition, our analysis identified several other non-B DNA motifs as new factors that potentially influence lentivirus integration, including human immunodeficiency virus type-1 in human cells. Together, our data demonstrate a clinically favorable integration site profile in the murine brain and identify non-B DNA as a potential new host factor that influences lentiviral integration in murine and human cells. PMID:25158091
PLoS One. 2014 Jul 1;9(7):e101436. doi: 10.1371/journal.pone.0101436. eCollection 2014.AltmetricsIn silico analysis of functional single nucleotide polymorphisms in the human TRIM22 gene.
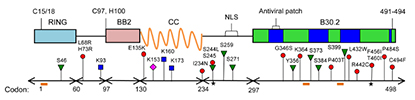
Abstract
Tripartite motif protein 22 (TRIM22) is an evolutionarily ancient protein that plays an integral role in the host innate immune response to viruses. The antiviral TRIM22 protein has been shown to inhibit the replication of a number of viruses, including HIV-1, hepatitis B, and influenza A. TRIM22 expression has also been associated with multiple sclerosis, cancer, and autoimmune disease. In this study, multiple in silico computational methods were used to identify non-synonymous or amino acid-changing SNPs (nsSNP) that are deleterious to TRIM22 structure and/or function. A sequence homology-based approach was adopted for screening nsSNPs in TRIM22, including six different in silico prediction algorithms and evolutionary conservation data from the ConSurf web server. In total, 14 high-risk nsSNPs were identified in TRIM22, most of which are located in a protein interaction module called the B30.2 domain. Additionally, 9 of the top high-risk nsSNPs altered the putative structure of TRIM22's B30.2 domain, particularly in the surface-exposed v2 and v3 regions. These same regions are critical for retroviral restriction by the closely-related TRIM5α protein. A number of putative structural and functional residues, including several sites that undergo post-translational modification, were also identified in TRIM22. This study is the first extensive in silico analysis of the highly polymorphic TRIM22 gene and will be a valuable resource for future targeted mechanistic and population-based studies. PMID:24983760
Hum Mutat. 2014 Sep;35(9):1072-81. doi: 10.1002/humu.22595. Epub 2014 Jun 24.AltmetricsAncient and recent adaptive evolution in the antiviral TRIM22 gene: identification of a single-nucleotide polymorphism that impacts TRIM22 function.
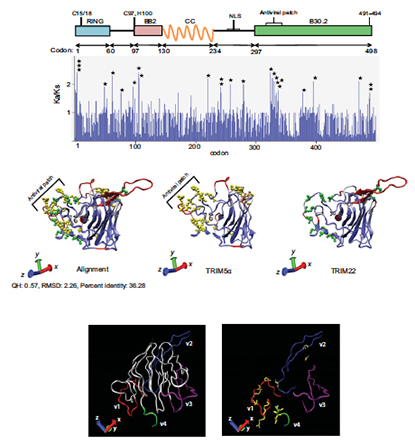
Abstract
Tripartite motif protein 22 (TRIM22) is a novel interferon-induced protein that potently inhibits the replication of evolutionarily diverse viruses, including HIV-1. Altered TRIM22 expression is also associated with diseases, such as multiple sclerosis, cancer, and autoimmunity. The factors that influence TRIM22 expression and antiviral activity are largely unknown. In this study, we adopted an evolution-guided functional approach to identify potential genetic determinants of TRIM22 function. Evolutionary analysis of TRIM22 from mammals spanning >100 million years demonstrated that TRIM22 evolution has been shaped by ancient and variable positive selection. We showed that positive selection is operating on multiple TRIM22 residues that cluster in putative functional regions and that some are predicted to be functionally damaging. Interestingly, the second most prevalent TRIM22 SNP in humans (rs1063303) is located at one of these positively selected sites. We showed that the frequency of rs1063303:G>C varies up to 10-fold between ethnicities and that in some ethnicities SNP rs1063303:G>C is being actively maintained in the population. The SNP rs1063303:G>C variant also had an inverse functional impact where it increased TRIM22 expression and decreased the antiviral activity of TRIM22. Taken together, our data characterize the extensive genetic variation in TRIM22 and identify rs1063303:G>C as a highly prevalent SNP that influences its function. PMID:24863734
Retrovirology. 2014 Apr 3;11:27. doi: 10.1186/1742-4690-11-27.AltmetricsInterferon-induced HERC5 is evolving under positive selection and inhibits HIV-1 particle production by a novel mechanism targeting Rev/RRE-dependent RNA nuclear export.
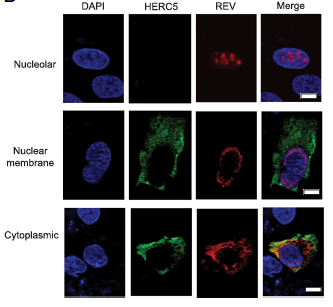
Abstract
BACKGROUND:
Type I interferon (IFN) inhibits virus replication by activating multiple antiviral mechanisms and pathways. It has long been recognized that type I IFNs can potently block HIV-1 replication in vitro; as such, HIV-1 has been used as a system to identify and characterize IFN-induced antiviral proteins responsible for this block. IFN-induced HERC5 contains an amino-terminal Regulator of Chromosome Condensation 1 (RCC1)-like domain and a carboxyl-terminal Homologous to the E6-AP Carboxyl Terminus (HECT) domain. HERC5 is the main cellular E3 ligase that conjugates the IFN-induced protein ISG15 to proteins. This E3 ligase activity was previously shown to inhibit the replication of evolutionarily diverse viruses, including HIV-1. The contribution of the RCC1-like domain to the antiviral activity of HERC5 was previously unknown. RESULTS:
In this study, we showed that HERC5 inhibits HIV-1 particle production by a second distinct mechanism that targets the nuclear export of Rev/RRE-dependent RNA. Unexpectedly, the E3 ligase activity of HERC5 was not required for this inhibition. Instead, this activity required the amino-terminal RCC1-like domain of HERC5. Inhibition correlated with a reduction in intracellular RanGTP protein levels and/or the ability of RanGTP to interact with RanBP1. Inhibition also correlated with altered subcellular localization of HIV-1 Rev. In addition, we demonstrated that positive evolutionary selection is operating on HERC5. We identified a region in the RCC1-like domain that exhibits an exceptionally high probability of having evolved under positive selection and showed that this region is required for HERC5-mediated inhibition of nuclear export. CONCLUSIONS:
We have identified a second distinct mechanism by which HERC5 inhibits HIV-1 replication and demonstrate that HERC5 is evolving under strong positive selection. Together, our findings contribute to a growing body of evidence suggesting that HERC5 is a novel host restriction factor. PMID: 24693865
Mol Biol Int. 2012;2012:153415. doi: 10.1155/2012/153415. Epub 2012 May 8.TRIM22: A Diverse and Dynamic Antiviral Protein.
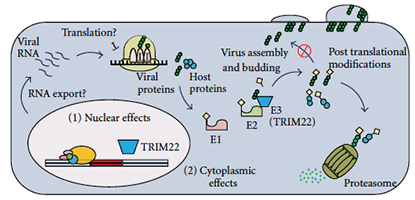
Abstract
The tripartite motif (TRIM) family of proteins is an evolutionarily ancient group of proteins with homologues identified in both invertebrate and vertebrate species. Human TRIM22 is one such protein that has a dynamic evolutionary history that includes gene expansion, gene loss, and strong signatures of positive selection. To date, TRIM22 has been shown to restrict the replication of a number of viruses, including encephalomyocarditis virus (EMCV), hepatitis B virus (HBV), and human immunodeficiency virus type 1 (HIV-1). In addition, TRIM22 has also been implicated in cellular differentiation and proliferation and may play a role in certain cancers and autoimmune diseases. This comprehensive paper summarizes our current understanding of TRIM22 structure and function. PMID: 22649727
Antiviral Res. 2012 Jan;93(1):175-84. doi: 10.1016/j.antiviral.2011.11.009. Epub 2011 Nov 26.Inhibition or deficiency of cathepsin B leads defects in HIV-1 Gag pseudoparticle release in macrophages and HEK293T cells.
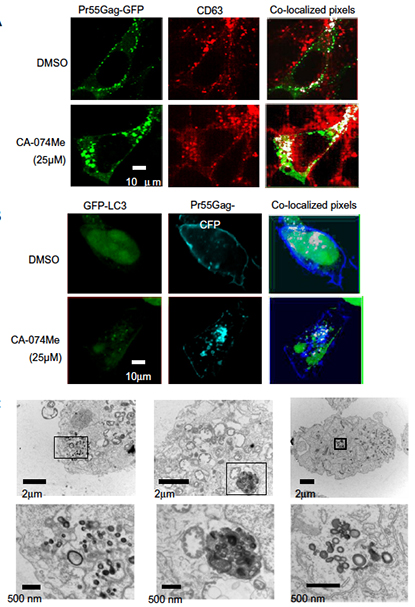
Abstract
Human immunodeficiency virus type 1 (HIV-1) egresses from infected cells through utilizing the host membrane budding mechanisms. Assembly of HIV-1 Gag particles occurs on membranes where the Gag multimers subsequently bud off and form enveloped viral particles. In certain cell types such as macrophages, HIV-1 Gag particles have shown to be released into intracellular virus containing compartments (VCC) such as late endosomes, multivesicular bodies (MVBs) or invaginated plasma membrane pockets. Here, we showed that macrophages or HEK293T cells treated with the cathepsin B (CTSB)-specific inhibitor CA-074Me or cells deficient in CTSB failed to release HIV-1 Gag pseudoparticles into the extracellular environment. Based on immunofluorescence and electron microscopy, these cells retained the pseudoparticles in heterogeneous intracellular VCC. CA-074Me was also able to inhibit propagation of two enveloped viruses, herpes simplex virus and influenza A virus, but not non-enveloped enterovirus. These results suggest that CTSB is required for the efficient release of HIV-1 Gag pseudoparticles and targeting CTSB can be a new therapeutic strategy for inhibiting egress of HIV-1 and other enveloped viruses. PMID: 22138708
Retrovirology. 2011 Nov 17;8:95. doi: 10.1186/1742-4690-8-95.AltmetricsHuman HERC5 restricts an early stage of HIV-1 assembly by a mechanism correlating with the ISGylation of Gag.
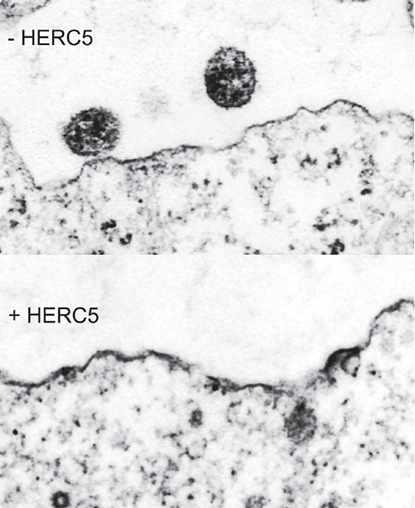
Abstract
BACKGROUND:
The identification and characterization of several interferon (IFN)-induced cellular HIV-1 restriction factors, defined as host cellular proteins or factors that restrict or inhibit the HIV-1 life cycle, have provided insight into the IFN response towards HIV-1 infection and identified new therapeutic targets for HIV-1 infection. To further characterize the mechanism underlying restriction of the late stages of HIV-1 replication, we assessed the ability of IFNbeta-induced genes to restrict HIV-1 Gag particle production and have identified a potentially novel host factor called HECT domain and RCC1-like domain-containing protein 5 (HERC5) that blocks a unique late stage of the HIV-1 life cycle. RESULTS:
HERC5 inhibited the replication of HIV-1 over multiple rounds of infection and was found to target a late stage of HIV-1 particle production. The E3 ligase activity of HERC5 was required for blocking HIV-1 Gag particle production and correlated with the post-translational modification of Gag with ISG15. HERC5 interacted with HIV-1 Gag and did not alter trafficking of HIV-1 Gag to the plasma membrane. Electron microscopy revealed that the assembly of HIV-1 Gag particles was arrested at the plasma membrane, at an early stage of assembly. The mechanism of HERC5-induced restriction of HIV-1 particle production is distinct from the mechanism underlying HIV-1 restriction by the expression of ISG15 alone, which acts at a later step in particle release. Moreover, HERC5 restricted murine leukemia virus (MLV) Gag particle production, showing that HERC5 is effective in restricting Gag particle production of an evolutionarily divergent retrovirus. CONCLUSIONS:
HERC5 represents a potential new host factor that blocks an early stage of retroviral Gag particle assembly. With no apparent HIV-1 protein that directly counteracts it, HERC5 may represent a new candidate for HIV/AIDS therapy. Comment in
- PMID: 22093708
-
PLoS Pathog. 2008 Feb 29;4(2):e1000007. doi: 10.1371/journal.ppat.1000007.Altmetrics
The interferon response inhibits HIV particle production by induction of TRIM22.
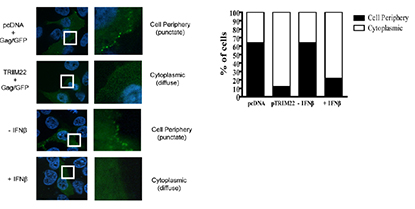
Abstract
Treatment of human cells with Type 1 interferons restricts HIV replication. Here we report that the tripartite motif protein TRIM22 is a key mediator. We used transcriptional profiling to identify cellular genes that were induced by interferon treatment and identified TRIM22 as one of the most strongly up-regulated genes. We confirmed, as in previous studies, that TRIM22 over-expression inhibited HIV replication. To assess the role of TRIM22 expressed under natural inducing conditions, we compared the effects of interferon in cells depleted for TRIM22 using RNAi and found that HIV particle release was significantly increased in the knockdown, implying that TRIM22 acts as a natural antiviral effector. Further studies showed that TRIM22 inhibited budding of virus-like particles containing Gag only, indicating that Gag was the target of TRIM22. TRIM22 did not block the release of MLV or EIAV Gag particles. Inhibition was associated with diffuse cytoplasmic staining of HIV Gag rather than accumulation at the plasma membrane, suggesting TRIM22 disrupts proper trafficking. Mutational analyses of TRIM22 showed that the catalytic amino acids Cys15 and Cys18 of the RING domain are required for TRIM22 antiviral activity. These data disclose a pathway by which Type 1 interferons obstruct HIV replication. PMID: 18389079-
Mol Ther. 2006 Aug;14(2):218-25. Epub 2006 May 2.
HIV integration site selection: targeting in macrophages and the effects of different routes of viral entry.
Abstract
We have studied the selection of HIV DNA integration sites in primary macrophages to investigate two questions. First, mature macrophages do not divide, allowing us to investigate whether HIV integration targeting differs between dividing cells and nondividing cells. We sequenced and analyzed 754 unique integration sites and found that integration in macrophages is favored in active transcription units (TUs), as was observed previously for other cell types. However, HIV integration in genes was slightly less favored in macrophages than in dividing PBMC or T cell lines. Second, we compared integration targeting by HIV-vector particles bearing either of two different envelope proteins (HIV R5 Env or VSV-G) to determine whether the mechanism of entry influenced subsequent integration targeting. Integration sites generated by HIV R5- or VSV-G-bearing particles showed no significant differences in their distributions in the human genome. Analysis of additional published integration site sequences also indicated that the route of entry did not affect integration site selection for other viral envelopes as well. PMID:16647883-
Nat Rev Microbiol. 2005 Nov;3(11):848-58.Altmetrics
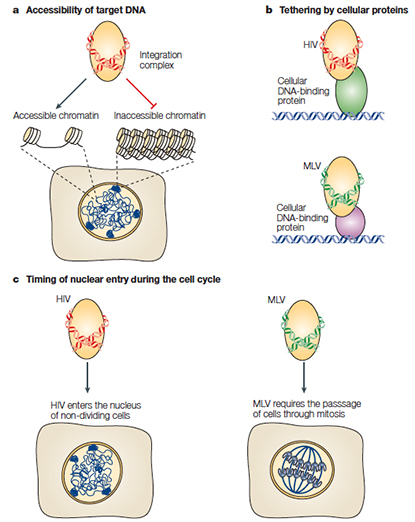
Genome-wide analysis of retroviral DNA integration.
Abstract
Retroviral vectors are often used to introduce therapeutic sequences into patients' cells. In recent years, gene therapy with retroviral vectors has had impressive therapeutic successes, but has also resulted in three cases of leukaemia caused by insertional mutagenesis, which has focused attention on the molecular determinants of retroviral-integration target-site selection. Here, we review retroviral DNA integration, with emphasis on recent genome-wide studies of targeting and on the status of efforts to modulate target-site selection. PMID:16175173
J Virol. 2005 Sep;79(18):12035-44.AltmetricsIntegration targeting by avian sarcoma-leukosis virus and human immunodeficiency virus in the chicken genome.

Abstract
We have analyzed the placement of sites of integration of avian sarcoma-leukosis virus (ASLV) and human immunodeficiency virus (HIV) DNA in the draft chicken genome sequence, with the goals of assessing species-specific effects on integration and allowing comparison to the distribution of chicken endogenous retroviruses (ERVs). We infected chicken embryo fibroblasts (CEF) with ASLV or HIV and sequenced 863 junctions between host and viral DNA. The relationship with cellular gene activity was analyzed by transcriptional profiling of uninfected or ASLV-infected CEF cells. ASLV weakly favored integration in active transcription units (TUs), and HIV strongly favored active TUs, trends seen previously for integration in human cells. The ERVs, in contrast, accumulated mostly outside TUs, including ERVs related to ASLV. The minority of ERVs present within TUs were mainly in the antisense orientation; consequently, the viral splicing and polyadenylation signals would not disrupt cellular mRNA synthesis. In contrast, de novo ASLV integration sites within TUs showed no orientation bias. Comparing the distribution of de novo ASLV integration sites to ERVs indicated that purifying selection against gene disruption, and not initial integration targeting, probably determined the ERV distribution. Further analysis indicated that ERVs in humans, mice, and rats showed similar distributions, suggesting purifying selection dictated their distributions as well. PMID:16140779
-