Tools
Access the application tool here
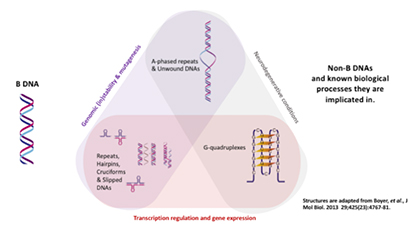
DNA was first identified in the late 1860s by Swiss chemist Friedrich Miescher. In the decades following, other researchers revealed additional details about the DNA molecule, including its primary chemical components and the ways in which they joined with one another. These laid the foundation for the ground breaking description of the DNA by Watson and Crick in 1953. It is now known that genomic DNA is not limited to the confines of the canonical B-forming DNA duplex described by Watson and Crick, but includes other different types of secondary structures collectively termed non-B DNA structures. Non-B DNA have recently garnered much interest because of their important roles in a variety of biological processes such as DNA replication, telomere maintenance, gene expression, viral replication, immune evasion, neurological disorders and cancers.
Non-B DNA Predictor identifies non-B DNA motifs in DNA (and RNA where applicable) sequences. Non-B DNA Predictor is based on gquad an R package, and such a valuable analytical tool for experimental biologists with little computational skills. The motifs that can be identified are A-phased DNA repeats, G-quadruplexes (with or without overlaps), intramolecular triplexes (with or without overlaps), slipped motifs, short tandem repeats, triplex forming oligonucleotides and Z-DNA motifs.